Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.)
- PMID: 39409596
- PMCID: PMC11479026
- DOI: 10.3390/plants13192726
Genome-Wide Identification and Expression Analysis of GST Genes during Light-Induced Anthocyanin Biosynthesis in Mango (Mangifera indica L.)
Abstract
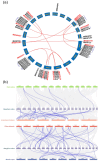
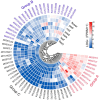
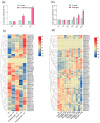
Anthocyanins are important secondary metabolites contributing to the red coloration of fruits, the biosynthesis of which is significantly affected by light. Glutathione S-transferases (GSTs) play critical roles in the transport of anthocyanins from the cytosol to the vacuole. Despite their importance, GST genes in mango have not been extensively characterized. In this study, 62 mango GST genes were identified and further divided into six subfamilies. MiGSTs displayed high similarity in their exon/intron structure and motif and domain composition within the same subfamilies. The mango genome harbored eleven pairs of segmental gene duplications and ten sets of tandemly duplicated genes. Orthologous analysis identified twenty-nine, seven, thirty-four, and nineteen pairs of orthologous genes among mango MiGST genes and their counterparts in Arabidopsis, rice, citrus, and bayberry, respectively. Tissue-specific expression profiling highlighted tissue-specific expression patterns for MiGST genes. RNA-seq and qPCR analyses revealed elevated expression levels of seven MiGSTs including MiDHAR1, MiGSTU7, MiGSTU13, MiGSTU21, MiGSTF3, MiGSTF8, and MiGSTF9 during light-induced anthocyanin accumulation in mango. This study establishes a comprehensive genetic framework of MiGSTs in mango fruit and their potential roles in regulating anthocyanin accumulation, which is helpful in developing GST-derived molecular markers and speeding up the process of breeding new red-colored mango cultivars.
Keywords: GST; anthocyanin; gene expression; light; mango.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Kanzaki S., Ichihi A., Tanaka Y., Fujishige S., Koeda S., Shimizu K. The R2R3-MYB Transcription Factor MiMYB1 Regulates Light Dependent Red Coloration of ‘Irwin’ Mango Fruit Skin. Sci. Hortic. 2020;272:109567. doi: 10.1016/j.scienta.2020.109567. - DOI
-
- Medlicott A.P., Bhogal M., Reynolds S.B. Changes in Peel Pigmentation during Ripening of Mango Fruit (Mangifera Indica Var. Tommy Atkins) Ann. Appl. Biol. 1986;109:651–656. doi: 10.1111/j.1744-7348.1986.tb03222.x. - DOI
-
- Shi B., Wu H., Zheng B., Qian M., Gao A., Zhou K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera Indica L.) Horticulturae. 2021;7:423. doi: 10.3390/horticulturae7110423. - DOI
-
- Li B., Zhang X., Duan R., Han C., Yang J., Wang L., Wang S., Su Y., Wang L., Dong Y., et al. Genomic Analysis of the Glutathione S-Transferase Family in Pear (Pyrus Communis) and Functional Identification of PcGST57 in Anthocyanin Accumulation. Int. J. Mol. Sci. 2022;23:746. doi: 10.3390/ijms23020746. - DOI - PMC - PubMed
-
- Zhang Y., Xu Y., Huang D., Xing W., Wu B., Wei Q., Xu Y., Zhan R., Ma F., Song S., et al. Research Progress on the MYB Transcription Factors in Tropical Fruit. Trop. Plants. 2022;1:5. doi: 10.48130/TP-2022-0005. - DOI
Grants and funding
- 32360736/National Natural Science Foundation of China
- 32160678/National Natural Science Foundation of China
- 2023YFD2300801/National Key Research and Development Plan of China
- ZDKJ2021014/Major Science and Technology Plan of Hainan Province
- XTCX2022NYC04/Collaborative Innovation Center of Nanfan and High-Efficiency Tropical Agriculture, Hainan University
LinkOut - more resources
Full Text Sources
Research Materials

