Genome-Wide Association Study of Reproductive Traits in Large White Pigs
- PMID: 39409823
- PMCID: PMC11475698
- DOI: 10.3390/ani14192874
Genome-Wide Association Study of Reproductive Traits in Large White Pigs
Abstract
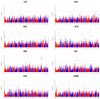
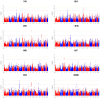
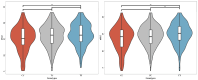
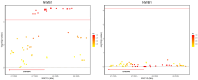
(1) Background: Reproductive performance is crucial for the pork industry's success. The Large White pig is central to this, yet the genetic factors influencing its reproductive traits are not well understood, highlighting the need for further research. (2) Methods: This study utilized Genome-Wide Association Studies to explore the genetic basis of reproductive traits in the Large White pig. We collected data from 2237 Large White sows across four breeding herds in southern China, focusing on eight reproductive traits. Statistical analyses included principal component analysis, linkage disequilibrium analysis, and univariate linear mixed models to identify significant single-nucleotide polymorphisms and candidate genes. (3) Results: Forty-five significantly related SNPs and 17 potential candidate genes associated with litter traits were identified. Individuals with the TT genotype at SNP rs341909772 showed an increase of 1.24 in the number of piglets born alive (NBA) and 1.25 in the number of healthy births (NHBs) compared with those with the CC genotype. (4) Conclusions: The SNPs and genes identified in this study offer insights into the genetics of reproductive traits in the Large White pig, potentially guiding the development of breeding strategies to improve litter size.
Keywords: GWAS; Large White pigs; SNPs; candidate genes; litter traits.
Conflict of interest statement
Authors Xiaoyan He, Dan Wu, and Yuxing Zhang were employed by Wens Foodstuff Group Co., Ltd. This company provided the pigs used in this study from a swine breeding herd that it owns and manages. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Knecht D., Środoń S., Duziński K. The impact of season, parity and breed on selected reproductive performance parameters of sows. Arch. Anim. Breed. 2015;58:49–56. doi: 10.5194/aab-58-49-2015. - DOI
-
- Zhang S., Zhang J., Olasege B.S., Ma P., Qiu X., Gao H., Wang C., Wang Y., Zhang Q., Yang H., et al. Estimation of genetic parameters for reproductive traits in connectedness groups of Duroc, Landrace and Yorkshire pigs in China. J. Anim. Breed. Genet. 2020;137:211–222. doi: 10.1111/jbg.12431. - DOI - PubMed
-
- Schiavo G., Bovo S., Tinarelli S., Gallo M., Dall’Olio S., Fontanesi L. Genome-wide association analyses for coat colour patterns in the autochthonous Nero Siciliano pig breed. Livest. Sci. 2020;236:104015. doi: 10.1016/j.livsci.2020.104015. - DOI
-
- Zhang Y., Zhang J., Gong H., Cui L., Zhang W., Ma J., Chen C., Ai H., Xiao S., Huang L., et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2018;150:47–55. doi: 10.1016/j.meatsci.2018.12.008. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

