Benchmarking of Approaches for Gene Copy-Number Variation Analysis and Its Utility for Genetic Aberration Detection in High-Grade Serous Ovarian Carcinomas
- PMID: 39409874
- PMCID: PMC11475927
- DOI: 10.3390/cancers16193252
Benchmarking of Approaches for Gene Copy-Number Variation Analysis and Its Utility for Genetic Aberration Detection in High-Grade Serous Ovarian Carcinomas
Abstract
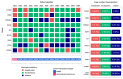
Objective: The goal of this study was to compare the results of CNV detection by three different methods using 13 paired carcinoma samples, as well as to perform a statistical analysis of the agreement. Methods: CNV was studied using NanoString nCounter v2 Cancer CN Assay (Nanostring), Illumina Infinium CoreExome microarrays (CoreExome microarrays) and digital droplet PCR (ddPCR). Results: There was a good level of agreement (PABAK score > 0.6) between the CoreExome microarrays and the ddPCR results for finding CNVs. There was a moderate level of agreement (PABAK values ≈ 0.3-0.6) between the NanoString Assay results and microarrays or ddPCR. For 83 out of 87 target genes studied (95%), the agreement between the CoreExome microarrays and NanoString nCounter was characterized by PABAK values < 0.75, except for MAGI3, PDGFRA, NKX2-1 and KDR genes (>0.75). The MET, HMGA2, KDR, C8orf4, PAX9, CDK6, and CCND2 genes had the highest agreement among all three approaches. Conclusions: Therefore, to get a better idea of how to genotype an unknown CNV spectrum in tumor or normal tissue samples that are very different molecularly, it makes sense to use at least two CNV detection methods. One of them, like ddPCR, should be able to quantitatively confirm the results of the other.
Keywords: CoreExome DNA microarray; HGSC; NanoString CNV panel; PABAK; benchmarking; digital droplet PCR (ddPCR); gene copy number variation; high-grade serous carcinoma; ovarian cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

