Dual Fractions Proteomic Analysis of Silica Nanoparticle Interactions with Protein Extracts
- PMID: 39410479
- PMCID: PMC11478063
- DOI: 10.3390/ma17194909
Dual Fractions Proteomic Analysis of Silica Nanoparticle Interactions with Protein Extracts
Abstract
Dual-fraction proteomics reveals a novel class of proteins impacted by nanoparticle exposure.
Background: Nanoparticles (NPs) interact with cellular proteomes, altering biological processes. Understanding these interactions requires comprehensive analyses beyond solely characterizing the NP corona.
Methods: We utilized a dual-fraction mass spectrometry (MS) approach to analyze both NP-bound and unbound proteins in Saccharomyces cerevisiae sp. protein extracts exposed to silica nanoparticles (SiNPs). We identified unique protein signatures for each fraction and quantified protein abundance changes using spectral counts.
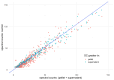
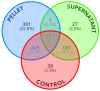
Results: Strong correlations were observed between protein profiles in each fraction and non-exposed controls, while minimal correlation existed between the fractions themselves. Linear models demonstrated equal contributions from both fractions in predicting control sample abundance. Combining both fractions revealed a larger proteomic response to SiNP exposure compared to single-fraction analysis. We identified 302/56 proteins bound/unbound to SiNPs and an additional 196 "impacted" proteins demonstrably affected by SiNPs.
Conclusion: This dual-fraction MS approach provides a more comprehensive understanding of nanoparticle interactions with cellular proteomes. It reveals a novel class of "impacted" proteins, potentially undergoing conformational changes or aggregation due to NP exposure. Further research is needed to elucidate their biological functions and the mechanisms underlying their impact.
Keywords: corona; mass spectrometry; protein extracts; protein–nanoparticle interactions; proteomics; silica nanoparticles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

