Virological characteristics of SARS-CoV-2 Omicron BA.5.2.48
- PMID: 39411709
- PMCID: PMC11473351
- DOI: 10.3389/fimmu.2024.1427284
Virological characteristics of SARS-CoV-2 Omicron BA.5.2.48
Abstract
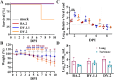
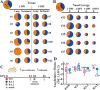
With the prevalence of sequentially-emerged sublineages including BA.1, BA.2 and BA.5, SARS-CoV-2 Omicron infection has transformed into a regional epidemic disease. As a sublineage of BA.5, the BA.5.2.48 outbroke and evolved into multi-subvariants in China without clearly established virological characteristics. Here, we evaluated the virological characteristics of two isolates of the prevalent BA.5.2.48 subvariant, DY.2 and DY.1.1 (a subvariant of DY.1). Compared to the normal BA.5 spike, the double-mutated DY.1.1 spike demonstrates efficient cleavage, reduced fusogenicity and higher hACE2 binding affinity. BA.5.2.48 demonstrated enhanced airborne transmission capacity than BA.2 in hamsters. The pathogenicity of BA.5.2.48 is greater than BA.2, as revealed in Omicron-lethal H11-K18-hACE2 rodents. In both naïve and convalescent hamsters, DY.1.1 shows stronger fitness than DY.2 in hamster turbinates. Thus regional outbreaking of BA.5.2.48 promotes the multidirectional evolution of its subvariants, gaining either enhanced pathogenicity or a fitness in upper airways which is associated with higher transmission.
Keywords: BA.5; Omicron; SARS-CoV-2; animal models; pathogenicity; spike.
Copyright © 2024 Wang, Jin, Liu, Zeng, Zhu, Li, Wang, Xiang, Zhang, Chen, Gao, Lai, Yan, Xia, Li, Wang and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

