Addressing the Generalizability of AI in Radiology Using a Novel Data Augmentation Framework with Synthetic Patient Image Data: Proof-of-Concept and External Validation for Classification Tasks in Multiple Sclerosis
- PMID: 39412405
- PMCID: PMC11605143
- DOI: 10.1148/ryai.230514
Addressing the Generalizability of AI in Radiology Using a Novel Data Augmentation Framework with Synthetic Patient Image Data: Proof-of-Concept and External Validation for Classification Tasks in Multiple Sclerosis
Abstract
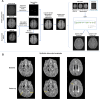
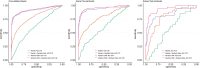
Artificial intelligence (AI) models often face performance drops after deployment to external datasets. This study evaluated the potential of a novel data augmentation framework based on generative adversarial networks (GANs) that creates synthetic patient image data for model training to improve model generalizability. Model development and external testing were performed for a given classification task, namely the detection of new fluid-attenuated inversion recovery lesions at MRI during longitudinal follow-up of patients with multiple sclerosis (MS). An internal dataset of 669 patients with MS (n = 3083 examinations) was used to develop an attention-based network, trained both with and without the inclusion of the GAN-based synthetic data augmentation framework. External testing was performed on 134 patients with MS from a different institution, with MR images acquired using different scanners and protocols than images used during training. Models trained using synthetic data augmentation showed a significant performance improvement when applied on external data (area under the receiver operating characteristic curve [AUC], 83.6% without synthetic data vs 93.3% with synthetic data augmentation; P = .03), achieving comparable results to the internal test set (AUC, 95.0%; P = .53), whereas models without synthetic data augmentation demonstrated a performance drop upon external testing (AUC, 93.8% on internal dataset vs 83.6% on external data; P = .03). Data augmentation with synthetic patient data substantially improved performance of AI models on unseen MRI data and may be extended to other clinical conditions or tasks to mitigate domain shift, limit class imbalance, and enhance the robustness of AI applications in medical imaging. Keywords: Brain, Brain Stem, Multiple Sclerosis, Synthetic Data Augmentation, Generative Adversarial Network Supplemental material is available for this article. © RSNA, 2024.
Keywords: Brain; Brain Stem; Generative Adversarial Network; Multiple Sclerosis; Synthetic Data Augmentation.
Conflict of interest statement
Figures
References
-
- Widner K , Virmani S , Krause J , et al. . Lessons learned from translating AI from development to deployment in healthcare . Nat Med 2023. ; 29 ( 6 ): 1304 – 1306 . - PubMed
-
- Rajpurkar P , Lungren MP . The Current and Future State of AI Interpretation of Medical Images . N Engl J Med 2023. ; 388 ( 21 ): 1981 – 1990 . - PubMed
-
- Domingos P . A few useful things to know about machine learning . Commun ACM 2012. ; 55 ( 10 ): 78 – 87 .
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

