A Plasmodium falciparum MORC protein complex modulates epigenetic control of gene expression through interaction with heterochromatin
- PMID: 39412522
- PMCID: PMC11483127
- DOI: 10.7554/eLife.92201
A Plasmodium falciparum MORC protein complex modulates epigenetic control of gene expression through interaction with heterochromatin
Abstract
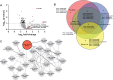
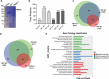
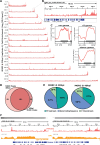
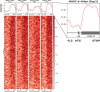
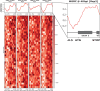
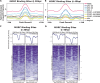
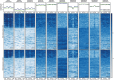
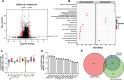
Dynamic control of gene expression is critical for blood stage development of malaria parasites. Here, we used multi-omic analyses to investigate transcriptional regulation by the chromatin-associated microrchidia protein, MORC, during asexual blood stage development of the human malaria parasite Plasmodium falciparum. We show that PfMORC (PF3D7_1468100) interacts with a suite of nuclear proteins, including APETALA2 (ApiAP2) transcription factors (PfAP2-G5, PfAP2-O5, PfAP2-I, PF3D7_0420300, PF3D7_0613800, PF3D7_1107800, and PF3D7_1239200), a DNA helicase DS60 (PF3D7_1227100), and other chromatin remodelers (PfCHD1 and PfEELM2). Transcriptomic analysis of PfMORCHA-glmS knockdown parasites revealed 163 differentially expressed genes belonging to hypervariable multigene families, along with upregulation of genes mostly involved in host cell invasion. In vivo genome-wide chromatin occupancy analysis during both trophozoite and schizont stages of development demonstrates that PfMORC is recruited to repressed, multigene families, including the var genes in subtelomeric chromosomal regions. Collectively, we find that PfMORC is found in chromatin complexes that play a role in the epigenetic control of asexual blood stage transcriptional regulation and chromatin organization.
Keywords: P. falciparum; PfMORC; Plasmodium falciparum; infectious disease; malaria; microbiology.
© 2023, Singh, Bonnell et al.
Conflict of interest statement
MS, VB, IT, VS, MM, JA, CD, GP, ML, CG No competing interests declared
Figures





Update of
- doi: 10.1101/2023.09.11.557196
- doi: 10.7554/eLife.92201.1
- doi: 10.7554/eLife.92201.2
References
-
- Antunes AV, Shahinas M, Swale C, Farhat DC, Ramakrishnan C, Bruley C, Cannella D, Robert MG, Corrao C, Couté Y, Hehl AB, Bougdour A, Coppens I, Hakimi M-A. In vitro production of cat-restricted Toxoplasma pre-sexual stages. Nature. 2024;625:366–376. doi: 10.1038/s41586-023-06821-y. - DOI - PMC - PubMed
-
- Arastu-Kapur S, Ponder EL, Fonović UP, Yeoh S, Yuan F, Fonović M, Grainger M, Phillips CI, Powers JC, Bogyo M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nature Chemical Biology. 2008;4:203–213. doi: 10.1038/nchembio.70. - DOI - PubMed
-
- Bonnell VA, Zhang Y, Brown JAS, Horton J, Josling GA, Chiu TP, Rohs R, Mahony S, Gordân R, Llinás M. DNA Sequence context and the chromatin landscape differentiate sequence-specific transcription factor binding in the human malaria parasite, Plasmodium falciparum. bioRxiv. 2023 doi: 10.1101/2023.03.31.535174. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

