Replication Stress Is an Actionable Genetic Vulnerability in Desmoplastic Small Round Cell Tumors
- PMID: 39412947
- PMCID: PMC11694066
- DOI: 10.1158/0008-5472.CAN-23-3603
Replication Stress Is an Actionable Genetic Vulnerability in Desmoplastic Small Round Cell Tumors
Abstract
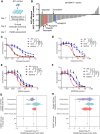
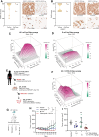
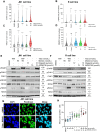
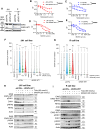
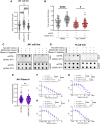
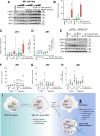
Desmoplastic small round cell tumor (DSRCT) is an aggressive sarcoma subtype that is driven by the EWS-WT1 chimeric transcription factor. The prognosis for DSRCT is poor, and major advances in treating DSRCT have not occurred for over two decades. To identify effective therapeutic approaches to target DSRCT, we conducted a high-throughput drug sensitivity screen in a DSRCT cell line assessing chemosensitivity profiles for 79 small-molecule inhibitors. DSRCT cells were sensitive to PARP inhibitors (PARPi) and ataxia-telangiectasia and Rad3-related inhibitors (ATRi), as monotherapies and in combination. These effects were recapitulated using multiple clinical PARPi and ATRi in three biologically distinct, clinically relevant models of DSRCT, including cell lines, a patient-derived xenograft-derived organoid model, and a cell line-derived xenograft mouse model. Mechanistically, exposure to a combination of PARPi and ATRi caused increased DNA damage, G2-M checkpoint activation, micronuclei accumulation, replication stress, and R-loop formation. EWS-WT1 silencing abrogated these phenotypes and was epistatic with exogenous expression of the R-loop resolution enzyme RNase H1 in reversing sensitivity to PARPi and ATRi monotherapies. The combination of PARPi and ATRi also induced EWS-WT1-dependent cell-autonomous activation of the cyclic GMP-AMP synthase-stimulator of IFN genes innate immune pathway and cell-surface expression of PD-L1. Taken together, these findings point toward a role for EWS-WT1 in generating R-loop-dependent replication stress that leads to a targetable vulnerability, providing a rationale for the clinical assessment of PARPi and ATRi in DSRCT. Significance: EWS-WT1, the unique oncogenic driver of desmoplastic small round cell tumors, confers sensitivity to PARP and ATR inhibitors, supporting the potential of these drugs in treating patients with this aggressive sarcoma subtype.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
M.M. Lenormand reports grants from Institut Pasteur outside the submitted work. N. Herbel reports personal fees from Viroxis and other support from Pharmaceutical Product Development and Thermo Fisher Scientific outside the submitted work. A. Loktev reports grants from The Kelly Turner Foundation during the conduct of the study. A. Zimmermann reports employment by Merck Healthcare KGaA, Darmstadt, Germany, during the conduct of the study. O. Mir reports personal fees from Amgen outside the submitted work. A. Le Cesne reports personal fees from PharmaMar and Deciphera outside the submitted work. M. Faron reports personal fees from Vifor Pharma outside the submitted work. C.J. Lord reports grants and personal fees from AstraZeneca, Merck KGaA, Artios, and NeoPhore; personal fees from FoRx, Syncona, Sun Pharma, Gerson Lehrman Group, Vertex, Third Rock, Ono Pharmaceutical, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, GlaxoSmithKline, Dawn Bioventures, Blacksmith Medicines, and Ariceum; personal fees and other support from Tango Therapeutics; and other support from OVIBIO and Hysplex during the conduct of the study, as well as being a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their development and use as part of the ICR's “Rewards to Inventors” scheme and also receiving benefits from this scheme associated with patents for PARP inhibitors paid into the author’s personal account and research accounts at the Institute of Cancer Research. S. Postel-Vinay reports research grants from AstraZeneca, Amgen, and Hoffman-La Roche/imCORE outside the submitted work. S. Postel-Vinay is also principal investigator of clinical trials sponsored by Amgen, Daiichi Sankyo, BeiGene, Bristol Myers Squibb, AstraZeneca, Clever Peptide, GlaxoSmithKline, Novartis, Oxford BioTherapeutics, and Roche (institutional funding for running the clinical trial); as well as personal fees from EPICS Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Figures
References
-
- Cidre-Aranaz F, Watson S, Amatruda JF, Nakamura T, Delattre O, de Alava E, et al. . Small round cell sarcomas. Nat Rev Dis Primer 2022;8:66. - PubMed
MeSH terms
Substances
Grants and funding
- ECO202206015528/Fondation pour la Recherche Médicale (FRM)
- European Society for Medical Oncology (ESMO)
- Canceropôle PACA (Canceropole PACA)
- Fondation Bettencourt Schueller (Bettencourt Schueller Foundation)
- Institut Servier (Servier Institute)
- Fondation des Treilles
- TargetSWitch 101077864/HORIZON EUROPE European Research Council (ERC)
- PGA1-RF20190208576/Fondation ARC pour la Recherche sur le Cancer (ARC)
- Fondation pour la Recherche Médicale (FRM)
- Institut National de la Santé et de la Recherche Médicale (Inserm)
- INCa-DGOS-Inserm_12551 SIRIC2 and INCa-DGOS-Inserm-ITMO Cancer_18002 SIRIC EpiCURE/Institut National Du Cancer (INCa)
- Recherche et Innovation en Cancé/Canceropôle PACA (Canceropole PACA)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

