Swine NONO promotes IRF3-mediated antiviral immune response by Detecting PRRSV N protein
- PMID: 39413144
- PMCID: PMC11482726
- DOI: 10.1371/journal.ppat.1012622
Swine NONO promotes IRF3-mediated antiviral immune response by Detecting PRRSV N protein
Abstract
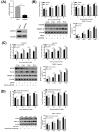
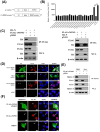
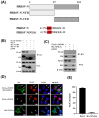
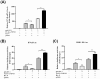
Non-POU domain-containing octamer-binding protein (NONO) is a multi-functional nuclear protein which belongs to the Drosophila behavior/human splicing (DBHS) protein family. NONO is known to regulate multiple important biological processes including host antiviral immune response. However, whether NONO can inhibit porcine reproductive and respiratory syndrome virus (PRRSV) replication is less well understood. In this study, we demonstrated that swine NONO (sNONO) inhibited PRRSV replication, via increasing expression of IFN-β, whereas NONO knockdown or knockout in PAM-KNU cells was more susceptible to PRRSV infection. As an IRF3 positive regulation factor, NONO promoted IFN-β expression by enhancing activation of IRF3. During PRRSV infection, NONO further up-regulated IRF3-mediated IFN-β expression by interacting with PRRSV N protein. Mechanistically, NONO functioned as a scaffold protein to detect PRRSV N protein and formed N-NONO-IRF3 complex in the nucleus. Interestingly, it was found that the NONO protein reversed the inhibitory effect of PRRSV N protein on type I IFN signaling pathway. Taken together, our study provides a novel mechanism for NONO to increase the IRF3-mediated IFN-β activation by interacting with the viral N protein to inhibit PRRSV infection.
Copyright: © 2024 Jiang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Zhang A, Duan H, Zhao H, Liao H, Du Y, Li L, et al.. Interferon-Induced Transmembrane Protein 3 Is a Virus-Associated Protein Which Suppresses Porcine Reproductive and Respiratory Syndrome Virus Replication by Blocking Viral Membrane Fusion. J Virol. 2020; 94(24). doi: 10.1128/JVI.01350-20 . - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

