Small molecules from antibody pharmacophores (SMAbPs) as a hit identification workflow for immune checkpoints
- PMID: 39413175
- PMCID: PMC11482313
- DOI: 10.1126/sciadv.adq5540
Small molecules from antibody pharmacophores (SMAbPs) as a hit identification workflow for immune checkpoints
Abstract
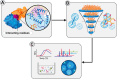
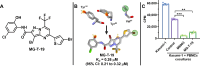
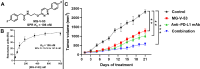
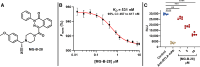
Small-molecule modulators of immune checkpoints are poised to revolutionize cancer immunotherapy. However, efficient strategies for hit identification are lacking. We introduce small molecules from antibody pharmacophores (SMAbPs), a workflow leveraging cocrystal structures of checkpoints with antibodies to create pharmacophore maps for virtual screening. Applying SMAbPs to five immune checkpoints yielded hits with submicromolar potency in both cell-free and cellular assays. Notably, SMAbPs identified the most potent T cell immunoglobulin and mucin-domain containing-3 and V-domain immunoglobulin suppressor of T cell activation (VISTA) inhibitors reported to date and first-in-class modulators of B and T lymphocyte attenuator, 4-IBB, and CD27. Targeting inhibitory and costimulatory checkpoints with hits identified through SMAbPs demonstrated remarkable in vivo antitumor activity, exemplified by MG-V-53 (VISTA inhibitor) and MG-C-30 (CD27 agonist), which significantly reduced tumor volumes in MC38 and EG7-OVA mouse models, respectively.
Figures
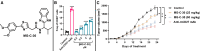
References
-
- Kruger S., Ilmer M., Kobold S., Cadilha B. L., Endres S., Ormanns S., Schuebbe G., Renz B. W., D’Haese J. G., Schloesser H., Heinemann V., Subklewe M., Boeck S., Werner J., Bergwelt-Baildon M., Advances in cancer immunotherapy 2019 – Latest trends. J. Exp. Clin. Cancer Res. 38, 268 (2019). - PMC - PubMed
-
- Wei S. C., Duffy C. R., Allison J. P., Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

