The clock-associated LUX ARRHYTHMO regulates high-affinity nitrate transport in Arabidopsis roots
- PMID: 39413246
- PMCID: PMC11629737
- DOI: 10.1111/tpj.17080
The clock-associated LUX ARRHYTHMO regulates high-affinity nitrate transport in Arabidopsis roots
Abstract
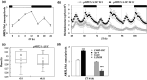
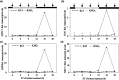
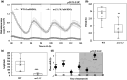
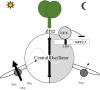
The circadian clock organizes physiological processes in plants to occur at specific times of the day, optimizing efficient use of resources. Nitrate is a crucial inorganic nitrogen source for agricultural systems to sustain crop productivity. However, because nitrate fertilization has a negative impact on the environment, it is important to carefully manage nitrate levels. Understanding crop biological rhythms can lead to more ecologically friendly agricultural practices. Gating responses through the circadian clock could be a strategy to enhance root nitrate uptake and to limit nitrate runoff. In Arabidopsis, the NITRATE TRANSPORTER 2.1 (NRT2.1) gene encodes a key component of the high-affinity nitrate transporter system. Our study reveals that NRT2.1 exhibits a rhythmic expression pattern, with daytime increases and nighttime decreases. The NRT2.1 promoter activity remains rhythmic under constant light, indicating a circadian regulation. The clock-associated transcription factor LUX ARRHYTHMO (LUX) binds to the NRT2.1 promoter in vivo. Loss-of-function of LUX leads to increased NRT2.1 transcript levels and root nitrate uptake at dusk. This supports LUX acting as a transcriptional repressor and modulating NRT2.1 expression in a time-dependent manner. Furthermore, applying nitrate at different times of the day results in varying magnitudes of the transcriptional response in nitrate-regulated genes. We also demonstrate that a defect in the high-affinity nitrate transport system feeds back to the central oscillator by modifying the LUX promoter activity. In conclusion, this study uncovers a molecular pathway connecting the root nitrate uptake and circadian clock, with potential agro-chronobiological applications.
Keywords: circadian clock; high‐affinity nitrate transport; nitrogen assimilation and metabolism; root uptake; transcription regulation.
© 2024 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Alvarez, J.M. , Riveras, E. , Vidal, E.A. , Gras, D.E. , Contreras‐López, O. , Tamayo, K.P. et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal, 80, 1–13. Available from: 10.1111/tpj.12618 - DOI - PubMed
-
- Bijay‐Singh, C.E. & Craswell, E. (2021) Fertilizers and nitrate pollution of surface and ground water: an increasingly pervasive global problem. SN Applied Sciences, 3, 518. Available from: 10.1007/s42452-021-04521-8 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

