Genetic architecture of routinely acquired blood tests in a British South Asian cohort
- PMID: 39414775
- PMCID: PMC11484750
- DOI: 10.1038/s41467-024-53091-x
Genetic architecture of routinely acquired blood tests in a British South Asian cohort
Abstract
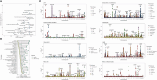
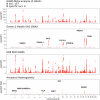
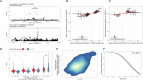
Understanding the genetic basis of routinely-acquired blood tests can provide insights into several aspects of human physiology. We report a genome-wide association study of 42 quantitative blood test traits defined using Electronic Healthcare Records (EHRs) of ~50,000 British Bangladeshi and British Pakistani adults. We demonstrate a causal variant within the PIEZO1 locus which was associated with alterations in red cell traits and glycated haemoglobin. Conditional analysis and within-ancestry fine mapping confirmed that this signal is driven by a missense variant - chr16-88716656-G-TT - which is common in South Asian ancestries (MAF 3.9%) but ultra-rare in other ancestries. Carriers of the T allele had lower mean HbA1c values, lower HbA1c values for a given level of random or fasting glucose, and delayed diagnosis of Type 2 Diabetes Mellitus. Our results shed light on the genetic basis of clinically-relevant traits in an under-represented population, and emphasise the importance of ancestral diversity in genetic studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

