Spatial proteomics identifies JAKi as treatment for a lethal skin disease
- PMID: 39415009
- PMCID: PMC11602713
- DOI: 10.1038/s41586-024-08061-0
Spatial proteomics identifies JAKi as treatment for a lethal skin disease
Abstract
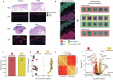
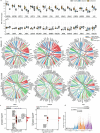
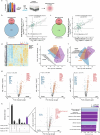
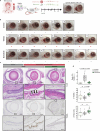
Toxic epidermal necrolysis (TEN) is a fatal drug-induced skin reaction triggered by common medications and is an emerging public health issue1-3. Patients with TEN undergo severe and sudden epidermal detachment caused by keratinocyte cell death. Although molecular mechanisms that drive keratinocyte cell death have been proposed, the main drivers remain unknown, and there is no effective therapy for TEN4-6. Here, to systematically map molecular changes that are associated with TEN and identify potential druggable targets, we utilized deep visual proteomics, which provides single-cell-based, cell-type-resolution proteomics7,8. We analysed formalin-fixed, paraffin-embedded archived skin tissue biopsies of three types of cutaneous drug reactions with varying severity and quantified more than 5,000 proteins in keratinocytes and skin-infiltrating immune cells. This revealed a marked enrichment of type I and type II interferon signatures in the immune cell and keratinocyte compartment of patients with TEN, as well as phosphorylated STAT1 activation. Targeted inhibition with the pan-JAK inhibitor tofacitinib in vitro reduced keratinocyte-directed cytotoxicity. In vivo oral administration of tofacitinib, baricitinib or the JAK1-specific inhibitors abrocitinib or upadacitinib ameliorated clinical and histological disease severity in two distinct mouse models of TEN. Crucially, treatment with JAK inhibitors (JAKi) was safe and associated with rapid cutaneous re-epithelialization and recovery in seven patients with TEN. This study uncovers the JAK/STAT and interferon signalling pathways as key pathogenic drivers of TEN and demonstrates the potential of targeted JAKi as a curative therapy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A patent for treatment of TEN, SJS–TEN and SJS with JAKi has been filed (US patent: provisional application no. 63/531,677; European patent: provisional application no. 23 190 686.8, with T.M.N., L.E.F. and M.M. as inventors). The patent application belongs to the Max-Planck-Gesellschaft zur Förderung der Wissenschaften e.V. M.M. is an indirect investor in Evosep Biosystems. The authors declare no other competing interests.
Figures












Comment in
-
JAK inhibition combats adverse skin reaction.Nat Rev Drug Discov. 2024 Dec;23(12):897. doi: 10.1038/d41573-024-00181-6. Nat Rev Drug Discov. 2024. PMID: 39501097 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

