Circadian rhythms of macrophages are altered by the acidic tumor microenvironment
- PMID: 39415049
- PMCID: PMC11549407
- DOI: 10.1038/s44319-024-00288-2
Circadian rhythms of macrophages are altered by the acidic tumor microenvironment
Abstract
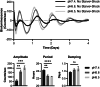
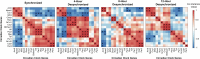
Tumor-associated macrophages (TAMs) are prime therapeutic targets due to their pro-tumorigenic functions, but varying efficacy of macrophage-targeting therapies highlights our incomplete understanding of how macrophages are regulated within the tumor microenvironment (TME). The circadian clock is a key regulator of macrophage function, but how circadian rhythms of macrophages are influenced by the TME remains unknown. Here, we show that conditions associated with the TME such as polarizing stimuli, acidic pH, and lactate can alter circadian rhythms in macrophages. While cyclic AMP (cAMP) has been reported to play a role in macrophage response to acidic pH, our results indicate pH-driven changes in circadian rhythms are not mediated solely by cAMP signaling. Remarkably, circadian disorder of TAMs was revealed by clock correlation distance analysis. Our data suggest that heterogeneity in circadian rhythms within the TAM population level may underlie this circadian disorder. Finally, we report that circadian regulation of macrophages suppresses tumor growth in a murine model of pancreatic cancer. Our work demonstrates a novel mechanism by which the TME influences macrophage biology through modulation of circadian rhythms.
Keywords: Circadian Rhythms; Immuno-oncology; Immunology; Macrophage; Tumor Microenvironment.
© 2024. The Author(s).
Conflict of interest statement
BJA is a member of
Figures





Update of
-
Circadian rhythms of macrophages are altered by the acidic pH of the tumor microenvironment.bioRxiv [Preprint]. 2024 Jul 24:2024.02.14.580339. doi: 10.1101/2024.02.14.580339. bioRxiv. 2024. Update in: EMBO Rep. 2024 Nov;25(11):5080-5112. doi: 10.1038/s44319-024-00288-2. PMID: 38405770 Free PMC article. Updated. Preprint.
References
-
- Adrover JM, Del Fresno C, Crainiciuc G, Cuartero MI, Casanova-Acebes M, Weiss LA, Huerga-Encabo H, Silvestre-Roig C, Rossaint J, Cossio I et al (2019) A neutrophil timer coordinates immune defense and vascular protection. Immunity 50:390–402.e310 - PubMed
-
- Allen JE (2023) IL-4 and IL-13: regulators and effectors of wound repair. Annu Rev Immunol 41:229–254 - PubMed
MeSH terms
Substances
Grants and funding
- T32GM135134/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- T32AI007285/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- No grant number/University of Rochester (UR)
- P30 ES001247/ES/NIEHS NIH HHS/United States
- T32 GM135134/GM/NIGMS NIH HHS/United States
- R01CA282225/HHS | NIH | National Cancer Institute (NCI)
- R01 CA282225/CA/NCI NIH HHS/United States
- R00 CA204593/CA/NCI NIH HHS/United States
- T32 AI007285/AI/NIAID NIH HHS/United States
- No grant number/UR | URMC | Wilmot Cancer Institute, University of Rochester Medical Center (WCI)
- R00CA204593/HHS | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

