Copper aggravated synaptic damage after traumatic brain injury by downregulating BNIP3-mediated mitophagy
- PMID: 39415457
- PMCID: PMC11849941
- DOI: 10.1080/15548627.2024.2409613
Copper aggravated synaptic damage after traumatic brain injury by downregulating BNIP3-mediated mitophagy
Abstract
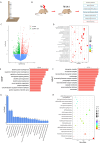
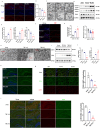
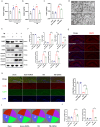
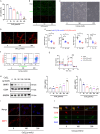
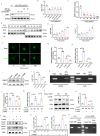
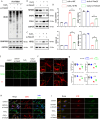
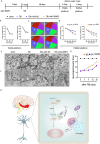
Synaptic damage is a crucial pathological process in traumatic brain injury. However, the mechanisms driving this process remain poorly understood. In this report, we demonstrate that the accumulation of damaged mitochondria, resulting from impaired mitphagy, plays a significant role in causing synaptic damage. Moreover, copper induced downregulation of BNIP3 is a key player in regulating mitophagy. DMSA alleviates synaptic damage and mitochondrial dysfunction by promoting urinary excretion of copper. Mechanistically, we find that copper downregulate BNIP3 by increasing the nuclear translocation of NFKB, which is triggered by TRIM25-mediated ubiquitination-dependent degradation of NFKBIA. Our study underscores the importance of copper accumulation in the regulation of BNIP3-mediated mitophagy and suggests that therapeutic targeting of the copper-TRIM25-NFKB-BNIP3 axis holds promise to attenuate synaptic damage after traumatic brain injury.
Keywords: BNIP3; TRIM25; copper; mitophagy; traumatic brain injury.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
