Small Molecule Screening Identifies HSP90 as a Modifier of RNA Foci in Myotonic Dystrophy Type 1
- PMID: 39415708
- PMCID: PMC12317676
- DOI: 10.1080/10985549.2024.2408025
Small Molecule Screening Identifies HSP90 as a Modifier of RNA Foci in Myotonic Dystrophy Type 1
Abstract
Myotonic dystrophy type 1 (DM1) is a multisystemic disorder caused by a CTG triplet repeat expansion within the 3' untranslated region of the DMPK gene. Expression of the expanded allele generates RNA containing long tracts of CUG repeats (CUGexp RNA) that form hairpin structures and accumulate in nuclear RNA foci; however, the factors that control DMPK expression and the formation of CUGexp RNA foci remain largely unknown. We performed an unbiased small molecule screen in an immortalized human DM1 skeletal muscle myoblast cell line and identified HSP90 as a modifier of endogenous RNA foci. Small molecule inhibition of HSP90 leads to enhancement of RNA foci and upregulation of DMPK mRNA levels. Knockdown and overexpression of HSP90 in undifferentiated DM1 myoblasts validated the impact of HSP90 with upregulation and downregulation of DMPK mRNA, respectively. Furthermore, we identified p-STAT3 as a downstream mediator of HSP90 impacting levels of DMPK mRNA and RNA foci. Interestingly, differentiated cells exhibited an opposite effect of HSP90 inhibition displaying downregulation of DMPK mRNA through a mechanism independent of p-STAT3 involvement. This study has revealed a novel mediator for DMPK mRNA and foci regulation in DM1 cells with the potential to identify targets for future therapeutic intervention.
Keywords: HSP90; Myotonic dystrophy; RNA foci; small molecule screening.
Conflict of interest statement
Disclosure Statement
The authors report there are no competing interests to declare.
Figures


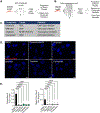

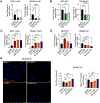


References
-
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Housman DE. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992; 68(4), 799–808. - PubMed
-
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O’hoy K, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992; 255(5049), 1253–1255. - PubMed
-
- Fu Y, Pizzuti A, Fenwick R Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, Jong PD, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science, 1992; 255(5049), 1256–1258. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
