This is a preprint.
Mesoscale regulation of MTOCs by the E3 ligase TRIM37
- PMID: 39416078
- PMCID: PMC11482927
- DOI: 10.1101/2024.10.09.617407
Mesoscale regulation of MTOCs by the E3 ligase TRIM37
Update in
-
Mesoscale regulation of microtubule-organizing centers by the E3 ligase TRIM37.Nat Struct Mol Biol. 2025 Sep;32(9):1787-1799. doi: 10.1038/s41594-025-01540-6. Epub 2025 May 25. Nat Struct Mol Biol. 2025. PMID: 40415023
Abstract
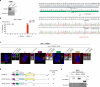
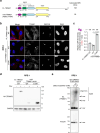
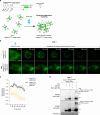
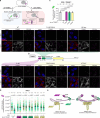
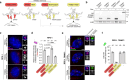
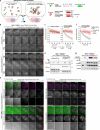
Centrosomes ensure accurate chromosome segregation during cell division. Although the regulation of centrosome number is well-established, less is known about the suppression of non-centrosomal MTOCs (ncMTOCs). The E3 ligase TRIM37, implicated in Mulibrey nanism and 17q23-amplified cancers, has emerged as a key regulator of both centrosomes and ncMTOCs. Yet, the mechanism by which TRIM37 achieves enzymatic activation to target these mesoscale structures had remained unknown. Here, we elucidate TRIM37's activation process, beginning with TRAF domain-directed substrate recognition, progressing through B-box domain-mediated oligomerization, and culminating in RING domain dimerization. Using optogenetics, we demonstrate that TRIM37's E3 activity is directly coupled to the assembly state of its substrates, activating only when centrosomal proteins cluster into higher-order assemblies resembling MTOCs. This regulatory framework provides a mechanistic basis for understanding TRIM37-driven pathologies and, by echoing TRIM5's restriction of the HIV capsid, unveils a conserved activation blueprint among TRIM proteins for controlling mesoscale assembly turnover.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
