This is a preprint.
Synergistic Role of Amino Acids in Enhancing mTOR Activation Through Lysosome Positioning
- PMID: 39416115
- PMCID: PMC11482915
- DOI: 10.1101/2024.10.12.618047
Synergistic Role of Amino Acids in Enhancing mTOR Activation Through Lysosome Positioning
Abstract
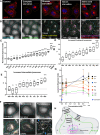
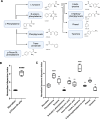
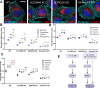
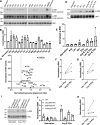
Lysosome positioning, or lysosome cellular distribution, is critical for lysosomal functions in response to both extracellular and intracellular cues. Amino acids, as essential nutrients, have been shown to promote lysosome movement toward the cell periphery. Peripheral lysosomes are involved in processes such as lysosomal exocytosis, cell migration, and metabolic signaling-functions that are particularly important for cancer cell motility and growth. However, the specific types of amino acids that regulate lysosome positioning, their underlying mechanisms, and their connection to amino acid-regulated metabolic signaling remain poorly understood. In this study, we developed a high-content imaging system for unbiased, quantitative analysis of lysosome positioning. We examined the 15 amino acids present in cell culture media and found that 10 promoted lysosome redistribution toward the cell periphery to varying extents, with aromatic amino acids showing the strongest effect. This redistribution was mediated by promoting outward transport through SLC38A9-BORC-kinesin 1/3 axis and simultaneously reducing inward transport via inhibiting the recruitment of Rab7 and JIP4 onto lysosomes. When examining the effects of amino acids on mTOR activation-a central regulator of cell metabolism-we found that the amino acids most strongly promoting lysosome dispersal, such as phenylalanine, did not activate mTOR on their own. However, combining phenylalanine with arginine, which activates mTOR without affecting lysosome positioning, synergistically enhanced mTOR activity. This synergy was lost when lysosomes failed to localize to the cell periphery, as observed in kinesin 1/3 knockout (KO) cells. Furthermore, breast cancer cells exhibited heightened sensitivity to phenylalanine-induced lysosome dispersal compared to noncancerous breast cells. Inhibition of LAT1, the amino acid transporter responsible for phenylalanine uptake, reduced peripheral lysosomes and impaired cancer cell migration and proliferation, highlighting the importance of lysosome positioning in these coordinated cellular activities. In summary, amino acid-regulated lysosome positioning and mTOR signaling depend on distinct sets of amino acids. Combining lysosome-dispersing amino acids with mTOR-activating amino acids synergistically enhances mTOR activation, which may be particularly relevant in cancer cells.
Keywords: High-content imaging; LAT1 (SLC7A5); amino acid signaling; bidirectional transport; breast cancer cells.
Figures


References
-
- Ballabio A., Bonifacino J.S., Lysosomes as dynamic regulators of cell and organismal homeostasis, Nat Rev Mol Cell Biol 21(2) (2020) 101–118. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
