This is a preprint.
Independent effects of testosterone, estradiol, and sex chromosomes on gene expression in immune cells of trans- and cisgender individuals
- PMID: 39416170
- PMCID: PMC11482753
- DOI: 10.1101/2024.10.08.617275
Independent effects of testosterone, estradiol, and sex chromosomes on gene expression in immune cells of trans- and cisgender individuals
Abstract
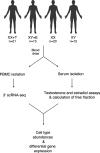
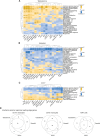
The origins of sex differences in human disease are elusive, in part because of difficulties in separating the effects of sex hormones and sex chromosomes. To separate these variables, we examined gene expression in four groups of trans- or cisgender individuals: XX individuals treated with exogenous testosterone (n=21), XY treated with exogenous estradiol (n=13), untreated XX (n=20), and untreated XY (n=15). We performed single-cell RNA-sequencing of 358,426 peripheral blood mononuclear cells. Across the autosomes, 8 genes responded with a significant change in expression to testosterone, 34 to estradiol, and 32 to sex chromosome complement with no overlap between the groups. No sex-chromosomal genes responded significantly to testosterone or estradiol, but X-linked genes responded to sex chromosome complement in a remarkably stable manner across cell types. Through leveraging a four-state study design, we successfully separated the independent actions of testosterone, estradiol, and sex chromosome complement on genome-wide gene expression in humans.
Keywords: Sex hormones; X chromosome; estradiol; gender; peripheral blood mononuclear cell; sex chromosomes; sex differences; single-cell RNA-sequencing; testosterone; transgender.
Conflict of interest statement
Declaration of interests: The authors declare no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
