This is a preprint.
Molecular model of a bacterial flagellar motor in situ reveals a "parts-list" of protein adaptations to increase torque
- PMID: 39416179
- PMCID: PMC11482838
- DOI: 10.1101/2023.09.08.556779
Molecular model of a bacterial flagellar motor in situ reveals a "parts-list" of protein adaptations to increase torque
Update in
-
In situ structure of a bacterial flagellar motor at subnanometre resolution reveals adaptations for increased torque.Nat Microbiol. 2025 Jul;10(7):1723-1740. doi: 10.1038/s41564-025-02012-9. Epub 2025 Jul 1. Nat Microbiol. 2025. PMID: 40595286 Free PMC article.
Abstract
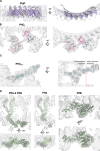
One hurdle to understanding how molecular machines work, and how they evolve, is our inability to see their structures in situ. Here we describe a minicell system that enables in situ cryogenic electron microscopy imaging and single particle analysis to investigate the structure of an iconic molecular machine, the bacterial flagellar motor, which spins a helical propeller for propulsion. We determine the structure of the high-torque Campylobacter jejuni motor in situ, including the subnanometre-resolution structure of the periplasmic scaffold, an adaptation essential to high torque. Our structure enables identification of new proteins, and interpretation with molecular models highlights origins of new components, reveals modifications of the conserved motor core, and explain how these structures both template a wider ring of motor proteins, and buttress the motor during swimming reversals. We also acquire insights into universal principles of flagellar torque generation. This approach is broadly applicable to other membrane-residing bacterial molecular machines complexes.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures











References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
