This is a preprint.
RPS19 and RPL5, the most commonly mutated genes in Diamond Blackfan anemia, impact DNA double-strand break repair
- PMID: 39416207
- PMCID: PMC11482920
- DOI: 10.1101/2024.10.10.617668
RPS19 and RPL5, the most commonly mutated genes in Diamond Blackfan anemia, impact DNA double-strand break repair
Abstract
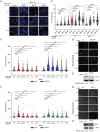
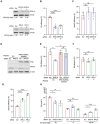
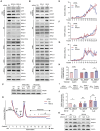
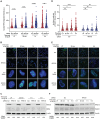
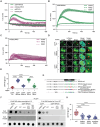
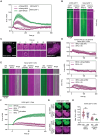
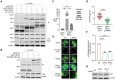
Diamond Blackfan anemia (DBA) is caused by germline heterozygous loss-of-function pathogenic variants (PVs) in ribosomal protein (RP) genes, most commonly RPS19 and RPL5. In addition to red cell aplasia, individuals with DBA are at increased risk of various cancers. Importantly, the mechanism(s) underlying cancer predisposition are poorly understood. We found that DBA patient-derived lymphoblastoid cells had persistent γ-H2AX foci following ionizing radiation (IR) treatment, suggesting DNA double-strand break (DSB) repair defects. RPS19- and RPL5-knocked down (KD) CD34+ cells had delayed repair of IR-induced DSBs, further implicating these RPs in DSB repair. Assessing the impact of RPS19- and RPL5-KD on specific DSB repair pathways, we found RPS19-KD decreased the efficiency of pathways requiring extensive end-resection, whereas RPL5-KD increased end-joining pathways. Additionally, RAD51 was reduced in RPS19- and RPL5-KD and RPS19- and RPL5-mutated DBA cells, whereas RPS19-deficient cells also had a reduction in PARP1 and BRCA2 proteins. RPS19-KD cells had an increase in nuclear RPA2 and a decrease in nuclear RAD51 foci post-IR, reflective of alterations in early, critical steps of homologous recombination. Notably, RPS19 and RPL5 interacted with poly(ADP)-ribose chains noncovalently, were recruited to DSBs in a poly(ADP)-ribose polymerase activity-dependent manner, and interacted with Ku70 and histone H2A. RPL5's recruitment, but not RPS19's, also required p53, suggesting that RPS19 and RPL5 directly participate in DSB repair via different pathways. We propose that defective DSB repair arising from haploinsufficiency of these RPs may underline the cancer predisposition in DBA.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
References
-
- Wlodarski MW, Vlachos A, Farrar JE, Da Costa LM, Kattamis A, Dianzani I, Belendez C, Unal S, Tamary H, Pasauliene R, Pospisilova D, de la Fuente J, Iskander D, Wolfe L, Liu JM, Shimamura A, Albrecht K, Lausen B, Bechensteen AG, Tedgard U, Puzik A, Quarello P, Ramenghi U, Bartels M, Hengartner H, Farah RA, Al Saleh M, Hamidieh AA, Yang W, Ito E, Kook H, Ovsyannikova G, Kager L, Gleizes PE, Dalle JH, Strahm B, Niemeyer CM, Lipton JM, Leblanc TM, international Diamond-Blackfan anaemia syndrome guideline p. Diagnosis, treatment, and surveillance of Diamond-Blackfan anaemia syndrome: international consensus statement. Lancet Haematol. 2024;11(5):e368–e382. - PubMed
-
- Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, Handsaker RE, Abdulhay NJ, Fiorini C, Genovese G, Lim ET, Cheng A, Cummings BB, Chao KR, Beggs AH, Genetti Ca, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Vlachos A, Lipton JM, Atsidaftos E, Glader B, Narla A, Gleizes PE, O'Donohue MF, Montel-Lehry N, Amor DJ, McCarroll SA, O'Donnell-Luria AH, Gupta N, Gabriel SB, MacArthur DG, Lander ES, Lek M, Da Costa L, Nathan DG, Korostelev AA, Do R, Sankaran VG, Gazda HT. The Genetic Landscape of Diamond-Blackfan Anemia. Am J Hum Genet. 2018;103(6):930–947. - PMC - PubMed
-
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schutz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA, Sankaran VG. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell. 2018;173(1):90–103 e119. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
