This is a preprint.
A noncanonical glycoprotein H complex enhances cytomegalovirus entry
- PMID: 39416215
- PMCID: PMC11482907
- DOI: 10.1101/2024.10.13.617647
A noncanonical glycoprotein H complex enhances cytomegalovirus entry
Update in
-
The GATE glycoprotein complex enhances human cytomegalovirus entry in endothelial cells.Nat Microbiol. 2025 Jul;10(7):1605-1616. doi: 10.1038/s41564-025-02025-4. Epub 2025 Jun 30. Nat Microbiol. 2025. PMID: 40588592 Free PMC article.
Abstract
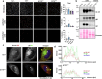
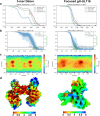
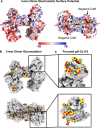
Human cytomegalovirus (HCMV) causes severe birth defects, lifelong health complications, and $4 billion in annual costs in the United States alone. A major challenge in vaccine design is the incomplete understanding of the diverse protein complexes the virus uses to infect cells. In Herpesviridae, the gH/gL glycoprotein heterodimer is expected to be a basal element of virion cell entry machinery. For HCMV, gH/gL forms a "trimer" with gO and a "pentamer" with UL128, UL130, and UL131A, with each complex binding distinct receptors to enter varied cell types. Here, we reveal a third glycoprotein complex, abundant in HCMV virions, which significantly enhances infection of endothelial cells. In this "3-mer" complex, gH, without gL, associates with UL116 and UL141, an immunoevasin previously known to function in an intracellular role. Cryo-EM reveals the virion-surface 3-mer is structurally unique among Herpesviridae gH complexes, with gH-only scaffolding, UL141-mediated dimerization and a heavily glycosylated UL116 cap. Given that antibodies directed at gH and UL141 each can restrict HCMV replication, our work highlights this virion surface complex as a new target for vaccines and antiviral therapies.
Conflict of interest statement
Competing interests. The authors declare no competing interests.
Figures
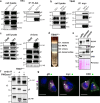
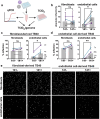
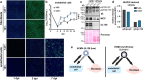
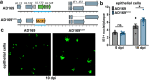






References
-
- Bossen C., Ingold K., Tardivel A., Bodmer J.-L., Gaide O., Hertig S., Ambrose C., Tschopp J., Schneider P., 2006. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J. Biol. Chem. 281, 13964–13971. - PubMed
-
- Caló S., Cortese M., Ciferri C., Bruno L., Gerrein R., Benucci B., Monda G., Gentile M., Kessler T., Uematsu Y., Maione D., Lilja A.E., Carfí A., Merola M., 2016. The Human Cytomegalovirus UL116 Gene Encodes an Envelope Glycoprotein Forming a Complex with gH Independently from gL. J. Virol. 90, 4926–4938. - PMC - PubMed
-
- Caposio P., van den Worm S., Crawford L., Perez W., Kreklywich C., Gilbride R.M., Hughes C.M., Ventura A.B., Ratts R., Marshall E.E., Malouli D., Axthelm M.K., Streblow D., Nelson J.A., Picker L.J., Hansen S.G., Früh K., 2019. Characterization of a live-attenuated HCMV-based vaccine platform. Sci. Rep. 9, 19236. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
