Single-cell transcriptional atlas of human breast cancers and model systems
- PMID: 39417215
- PMCID: PMC11483560
- DOI: 10.1002/ctm2.70044
Single-cell transcriptional atlas of human breast cancers and model systems
Abstract
Background: Breast cancer's complex transcriptional landscape requires an improved understanding of cellular diversity to identify effective treatments. The study of genetic variations among breast cancer subtypes at single-cell resolution has potential to deepen our insights into cancer progression.
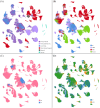
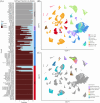
Methods: In this study, we amalgamate single-cell RNA sequencing data from patient tumours and matched lymph metastasis, reduction mammoplasties, breast cancer patient-derived xenografts (PDXs), PDX-derived organoids (PDXOs), and cell lines resulting in a diverse dataset of 117 samples with 506 719 total cells. These samples encompass hormone receptor positive (HR+), human epidermal growth factor receptor 2 positive (HER2+), and triple-negative breast cancer (TNBC) subtypes, including isogenic model pairs. Herein, we delineated similarities and distinctions across models and patient samples and explore therapeutic drug efficacy based on subtype proportions.
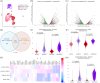
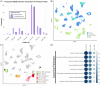
Results: PDX models more closely resemble patient samples in terms of tumour heterogeneity and cell cycle characteristics when compared with TNBC cell lines. Acquired drug resistance was associated with an increase in basal-like cell proportions within TNBC PDX tumours as defined with SCSubtype and TNBCtype cell typing predictors. All patient samples contained a mixture of subtypes; compared to primary tumours HR+ lymph node metastases had lower proportions of HER2-Enriched cells. PDXOs exhibited differences in metabolic-related transcripts compared to PDX tumours. Correlative analyses of cytotoxic drugs on PDX cells identified therapeutic efficacy was based on subtype proportion.
Conclusions: We present a substantial multimodel dataset, a dynamic approach to cell-wise sample annotation, and a comprehensive interrogation of models within systems of human breast cancer. This analysis and reference will facilitate informed decision-making in preclinical research and therapeutic development through its elucidation of model limitations, subtype-specific insights and novel targetable pathways.
Key points: Patient-derived xenografts models more closely resemble patient samples in tumour heterogeneity and cell cycle characteristics when compared with cell lines. 3D organoid models exhibit differences in metabolic profiles compared to their in vivo counterparts. A valuable multimodel reference dataset that can be useful in elucidating model differences and novel targetable pathways.
Keywords: breast cancer; cellular heterogeneity; model limitations; preclinical research; single‐cell RNA sequencing; single‐cell transcriptomics; subtype‐specific insights; targetable pathways; therapeutic drug efficacy.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The following authors disclosed conflicts of interest. Charles M. Perou is listed as an inventor on patent applications on the Breast PAM50 assay and is an equity stock holder and consultant of BioClassifier LLC. BDL and XSC are inventors (US Patent No. 11788147) of intellectual property (TNBCtype) licensed by Oncocyte Corp; the licensed IP is indirectly related to the work.
Figures





References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer J Clin. 2023;73:17‐48. - PubMed
-
- Desantis CE, Ma J, Gaudet MM. Breast cancer statistics, 2019. Cancer J Clin. 2019;69:438‐451. - PubMed
-
- Ma J, Jemal A. Breast Cancer Metastasis and Drug Resistance: Progress and Prospects. Springer; 2013:1‐18. doi:10.1007/978-1-4614-5647-6_1. Breast Cancer Statistics. ed. Ahmad, A.. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
