Genomic Landscape of ctDNA and Real-World Outcomes in Advanced Endometrial Cancer
- PMID: 39417689
- PMCID: PMC11647206
- DOI: 10.1158/1078-0432.CCR-24-2105
Genomic Landscape of ctDNA and Real-World Outcomes in Advanced Endometrial Cancer
Abstract
Purpose: ctDNA is a novel technique extensively studied in solid tumors, although not currently well defined in endometrial cancer.
Experimental design: A de-identified retrospective analysis of 1,988 patients with advanced/recurrent endometrial cancer was performed. In addition, an analysis of a real-world evidence cohort was completed (n = 1,266). Patients underwent ctDNA testing using Guardant360 during routine clinical care. The objective was to describe and assess molecular landscape using ctDNA.
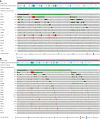
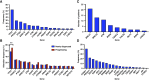
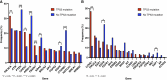
Results: Among 1,988 ctDNA samples, at least one somatic alteration was detected in 91.6% (n = 1,821). Most frequently altered genes were TP53 (64%), PIK3CA (29%), PTEN (25%), ARID1A (20%), and KRAS (14%). Overall, 18.5% had amplifications, with the majority identified in CCNE1 (40.9%), PIK3CA (22%), and EGFR (19.3%). From the real-world evidence cohort, those with TP53 mutations had a worse overall survival (OS) versus those without TP53 mutations (P = 0.02) and those with TP53 comutations had an inferior OS in comparison with TP53-mutated only (P = 0.016). Amongst these, patients with a PIK3CA comutation (P = 0.012) and CCNE1 amplification (P = 0.01) had an inferior OS compared with those with only TP53 mutations. Fifty-seven patients with newly diagnosed endometrial cancer had at least two serial ctDNA samples showing evolution in detected variants compared with baseline samples, with TP53 being the most frequent change.
Conclusions: This study is one of the largest cohorts of ctDNA currently reported in endometrial cancer. The presence of TP53 mutation and other comutations detected by ctDNA have a negative effect on outcomes. This report suggests that ctDNA analysis is feasible and could become a useful biomarker for endometrial cancer.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
K. Clemens reports being an employee and shareholder at Guardant Health. A. Bubie reports being an employee of Guardant Health, a liquid biopsy company focused on cancer detection. The technologies of Guardant Health were used to generate the data that were presented as part of this study. N. Zhang reports being a full-time employee and shareholder of Guardant Health. L. Drusbosky reports other support from Guardant Health during the conduct of the study. S. Lheureux reports fees from AstraZeneca, GSK, Repare Therapeutics, Merck, Eisai, Zai Lab, Gilead, Seagen, Abbvie and Schrodinger outside the submitted work. No disclosures were reported by the other authors.
Figures

References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7–33. - PubMed
-
- Tronconi F, Nero C, Giudice E, Salutari V, Musacchio L, Ricci C, et al. . Advanced and recurrent endometrial cancer: state of the art and future perspectives. Crit Rev Oncol Hematol 2022;180:103851. - PubMed
-
- Jamieson A, Barroilhet LM, McAlpine JN. Molecular classification in endometrial cancer: opportunities for precision oncology in a changing landscape. Cancer 2022;128:2853–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

