Transcriptome-Optimized Hydrogel Design of a Stem Cell Niche for Enhanced Tendon Regeneration
- PMID: 39417770
- PMCID: PMC11733723
- DOI: 10.1002/adma.202313722
Transcriptome-Optimized Hydrogel Design of a Stem Cell Niche for Enhanced Tendon Regeneration
Abstract
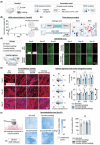
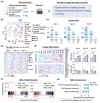
Bioactive hydrogels have emerged as promising artificial niches for enhancing stem cell-mediated tendon repair. However, a substantial knowledge gap remains regarding the optimal combination of niche features for targeted cellular responses, which often leads to lengthy development cycles and uncontrolled healing outcomes. To address this critical gap, an innovative, data-driven materiomics strategy is developed. This approach is based on in-house RNA-seq data that integrates bioinformatics and mathematical modeling, which is a significant departure from traditional trial-and-error methods. It aims to provide both mechanistic insights and quantitative assessments and predictions of the tenogenic effects of adipose-derived stem cells induced by systematically modulated features of a tendon-mimetic hydrogel (TenoGel). The knowledge generated has enabled a rational approach for TenoGel design, addressing key considerations, such as tendon extracellular matrix concentration, uniaxial tensile loading, and in vitro pre-conditioning duration. Remarkably, our optimized TenoGel demonstrated robust tenogenesis in vitro and facilitated tendon regeneration while preventing undesired ectopic ossification in a rat tendon injury model. These findings shed light on the importance of tailoring hydrogel features for efficient tendon repair. They also highlight the tremendous potential of the innovative materiomics strategy as a powerful predictive and assessment tool in biomaterial development for regenerative medicine.
Keywords: extracellular matrix; hydrogel; materiomics; stem cell therapy; tendon regeneration.
© 2024 The Author(s). Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Sharma P., Maffulli N., J. Bone Jt. Surg. Am. Vol. 2005, 87, 187. - PubMed
-
- Millar N. L., Silbernagel K. G., Thorborg K., Kirwan P. D., Galatz L. M., Abrams G. D., Murrell G. A. C., McInnes I. B., Rodeo S. A., Nat. Rev. Dis. Primers 2021, 7, 1. - PubMed
-
- Chamberlain G., Fox J., Ashton B., Middleton J., Stem Cells 2007, 25, 2739. - PubMed
-
- Lui P. P., Cheuk Y. C., Lee Y. W., Chan K. M., J. Orthop. Res. 2012, 30, 37. - PubMed
MeSH terms
Substances
Grants and funding
- GRF14121121/Research Grants Council of Hong Kong SAR
- GRF14118620/Research Grants Council of Hong Kong SAR
- ECS24201720:/Research Grants Council of Hong Kong SAR
- GRF14213922/Research Grants Council of Hong Kong SAR
- N_CUHK409/23/National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) of Hong Kong Joint Research Scheme
LinkOut - more resources
Full Text Sources
Miscellaneous

