DAB2IP loss in luminal a breast cancer leads to NF-κB-associated aggressive oncogenic phenotypes
- PMID: 39418101
- PMCID: PMC11623953
- DOI: 10.1172/jci.insight.171705
DAB2IP loss in luminal a breast cancer leads to NF-κB-associated aggressive oncogenic phenotypes
Abstract
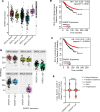
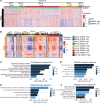
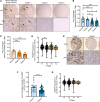
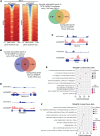
Despite proven therapy options for estrogen receptor-positive (ER+) breast tumors, a substantial number of patients with ER+ breast cancer exhibit relapse with associated metastasis. Loss of expression of RasGAPs leads to poor outcomes in several cancers, including breast cancer. Mining the The Cancer Genome Atlas (TCGA) breast cancer RNA-Seq dataset revealed that low expression of the RasGAP DAB2IP was associated with a significant decrease in relapse-free survival in patients with Luminal A breast cancer. Immunostaining demonstrated that DAB2IP loss occurred in grade 2 tumors and higher. Consistent with this, genes upregulated in DAB2IP-low Luminal A tumors were shared with more aggressive tumor subtypes and were associated with proliferation, metastasis, and altered ER signaling. Low DAB2IP expression in ER+ breast cancer cells was associated with increased proliferation, enhanced stemness phenotypes, and activation of IKK, the upstream regulator of the transcription factor NF-κB. Integrating cell-based ChIP-Seq with motif analysis and TCGA RNA-Seq data, we identified a set of candidate NF-κB target genes upregulated with loss of DAB2IP linked with several oncogenic phenotypes, including altered RNA processing. This study provides insight into mechanisms associated with aggressiveness and recurrence within a subset of the typically less aggressive Luminal A breast cancer intrinsic subtype.
Keywords: Breast cancer; NF-kappaB; Oncology; Tumor suppressors.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

