SUMOylation of Warts kinase promotes neural stem cell reactivation
- PMID: 39419973
- PMCID: PMC11487185
- DOI: 10.1038/s41467-024-52569-y
SUMOylation of Warts kinase promotes neural stem cell reactivation
Abstract
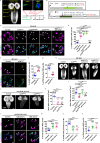
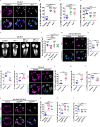
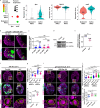
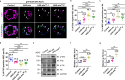
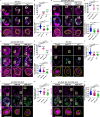
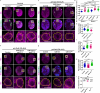
A delicate balance between neural stem cell (NSC) quiescence and proliferation is important for adult neurogenesis and homeostasis. Small ubiquitin-related modifier (SUMO)-dependent post-translational modifications cause rapid and reversible changes in protein functions. However, the role of the SUMO pathway during NSC reactivation and brain development is not established. Here, we show that the key components of the SUMO pathway play an important role in NSC reactivation and brain development in Drosophila. Depletion of SUMO/Smt3 or SUMO conjugating enzyme Ubc9 results in notable defects in NSC reactivation and brain development, while their overexpression leads to premature NSC reactivation. Smt3 protein levels increase with NSC reactivation, which is promoted by the Ser/Thr kinase Akt. Warts/Lats, the core protein kinase of the Hippo pathway, can undergo SUMO- and Ubc9-dependent SUMOylation at Lys766. This modification attenuates Wts phosphorylation by Hippo, leading to the inhibition of the Hippo pathway, and consequently, initiation of NSC reactivation. Moreover, inhibiting Hippo pathway effectively restores the NSC reactivation defects induced by SUMO pathway inhibition. Overall, our study uncovered an important role for the SUMO-Hippo pathway during Drosophila NSC reactivation and brain development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Morshead, C. M. et al. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron13, 1071–1082 (1994). - PubMed
-
- Tarpey, P. S. et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet.80, 345–352 (2007). - PMC - PubMed
-
- Badura-Stronka, M. et al. A novel nonsense mutation in CUL4B gene in three brothers with X-linked mental retardation syndrome. Clin. Genet.77, 141–144 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

