Macropinocytosis mediates resistance to loss of glutamine transport in triple-negative breast cancer
- PMID: 39420093
- PMCID: PMC11611898
- DOI: 10.1038/s44318-024-00271-6
Macropinocytosis mediates resistance to loss of glutamine transport in triple-negative breast cancer
Abstract
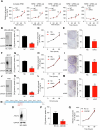
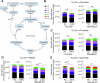
Triple-negative breast cancer (TNBC) metabolism and cell growth uniquely rely on glutamine uptake by the transporter ASCT2. Despite previous data reporting cell growth inhibition after ASCT2 knockdown, we here show that ASCT2 CRISPR knockout is tolerated by TNBC cell lines. Despite the loss of a glutamine transporter and low rate of glutamine uptake, intracellular glutamine steady-state levels were increased in ASCT2 knockout compared to control cells. Proteomics analysis revealed upregulation of macropinocytosis, reduction in glutamine efflux and increased glutamine synthesis in ASCT2 knockout cells. Deletion of ASCT2 in the TNBC cell line HCC1806 induced a strong increase in macropinocytosis across five ASCT2 knockout clones, compared to a modest increase in ASCT2 knockdown. In contrast, ASCT2 knockout impaired cell proliferation in the non-macropinocytic HCC1569 breast cancer cells. These data identify macropinocytosis as a critical secondary glutamine acquisition pathway in TNBC and a novel resistance mechanism to strategies targeting glutamine uptake alone. Despite this adaptation, TNBC cells continue to rely on glutamine metabolism for their growth, providing a rationale for targeting of more downstream glutamine metabolism components.
Keywords: ASCT2; Glutamine Metabolism; Macropinocytosis; Metabolomics; Triple-Negative Breast Cancer.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures









References
-
- Beckers RK, Selinger CI, Vilain R, Madore J, Wilmott JS, Harvey K, Holliday A, Cooper CL, Robbins E, Gillett D et al (2016) Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology 69:25–34 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

