RNA binding protein ZCCHC24 promotes tumorigenicity in triple-negative breast cancer
- PMID: 39420119
- PMCID: PMC11624195
- DOI: 10.1038/s44319-024-00282-8
RNA binding protein ZCCHC24 promotes tumorigenicity in triple-negative breast cancer
Abstract
Triple-negative breast cancer (TNBC) lacks the expression of hormone and HER2 receptors and is highly malignant with no effective therapeutic targets. In TNBC, the cancer stem-like cell (CSC) population is considered to be the main cause of resistance to treatment. Thus, the therapeutic targeting of this population could substantially improve patient survival. Here, we identify the RNA-binding protein ZCCHC24 as enriched in the mesenchymal-like TNBC population. ZCCHC24 promotes the expression of a set of genes related to tumorigenicity and treatment resistance by directly binding to the cis-element "UGUWHWWA" in their mRNAs, thereby stabilizing them. One of the ZCCHC24 targets, ZEB1, is a transcription factor that promotes the expression of cancer stemness genes and reciprocally induces ZCCHC24 expression. ZCCHC24 knockdown by siRNAs shows a therapeutic effect and reduces the mesenchymal-like cell population in TNBC patient-derived xenografts. ZCCHC24 knockdown also has additive effects with the BET inhibitor JQ1 in suppressing tumor growth in TNBC patient-derived xenografts.
Keywords: Breast Cancer; Cancer Stem Cells; RNA Binding Protein; ZEB1; mRNA Stabilization.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures



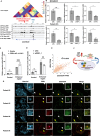




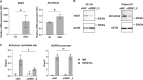

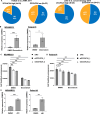
References
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457:608–611 - PubMed
-
- Batlle E, Clevers H (2017) Cancer stem cells revisited. Nat Med 23:1124–1134 - PubMed
MeSH terms
Substances
Grants and funding
- JP21K19403/MEXT | Japan Society for the Promotion of Science (JSPS)
- AR080127/HHS | National Institutes of Health (NIH)
- JP23gm0010009/Japan Agency for Medical Research and Development (AMED)
- R01 AR080127/AR/NIAMS NIH HHS/United States
- JP20H05696/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

