Feedback regulation of retinaldehyde reductase DHRS3, a critical determinant of retinoic acid homeostasis
- PMID: 39420244
- PMCID: PMC11808460
- DOI: 10.1002/1873-3468.15038
Feedback regulation of retinaldehyde reductase DHRS3, a critical determinant of retinoic acid homeostasis
Abstract
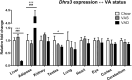
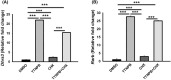
Retinoic acid is crucial for vertebrate embryogenesis, influencing anterior-posterior patterning and organogenesis through its interaction with nuclear hormone receptors comprising heterodimers of retinoic acid receptors (RARα, β, or γ) and retinoid X receptors (RXRα, β, or γ). Tissue retinoic acid levels are tightly regulated since both its excess and deficiency are deleterious. Dehydrogenase/reductase 3 (DHRS3) plays a critical role in this regulation by converting retinaldehyde to retinol, preventing excessive retinoic acid formation. Mutations in DHRS3 can result in embryonic lethality and congenital defects. This study shows that mouse Dhrs3 expression is responsive to vitamin A status and is directly regulated by the RAR/RXR complex through cis-regulatory elements. This highlights a negative feedback mechanism that ensures retinoic acid homeostasis.
Keywords: homeostasis; metabolism; negative feedback; nuclear hormone receptors; retinoic acid receptor; retinoids; transcriptional regulation; vitamin A.
© 2024 The Author(s). FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no commercial or financial relationships that could be construed as potential conflicts of interest regarding this research.
Figures


References
-
- Petkovich M, Brand NJ, Krust A and Chambon P (1987) A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature 330, 444–450. - PubMed
-
- Giguere V, Ong ES, Segui P and Evans RM (1987) Identification of a receptor for the morphogen retinoic acid. Nature 330, 624–629. - PubMed
-
- de The H, Vivanco‐Ruiz MM, Tiollais P, Stunnenberg H and Dejean A (1990) Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature 343, 177–180. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

