Dual diagnosis of UQCRFS1-related mitochondrial complex III deficiency and recessive GJA8-related cataracts
- PMID: 39421685
- PMCID: PMC11484756
- DOI: 10.1016/j.rare.2024.100040
Dual diagnosis of UQCRFS1-related mitochondrial complex III deficiency and recessive GJA8-related cataracts
Abstract
Biallelic pathogenic variants in UQCRFS1 underlie a rare form of isolated mitochondrial complex III deficiency associated with lactic acidosis and a distinctive scalp alopecia previously described in two unrelated probands. Here, we describe a participant in the Undiagnosed Diseases Network (UDN) with a dual diagnosis of two autosomal recessive disorders revealed by genome sequencing: UQCRFS1-related mitochondrial complex III deficiency and GJA8-related cataracts. Both pathogenic variants have been reported before: UQCRFS1 (NM_006003.3:c.215-1 G>C, p.Val72_Thr81del10) in a case with mitochondrial complex III deficiency and GJA8 (NM 005267.5:c.736 G>T, p.Glu246*) as a somatic change in aged cornea leading to decreased junctional coupling. A multi-modal approach combining enzyme assays and cellular proteomics analysis provided clear evidence of complex III respiratory chain dysfunction and low abundance of the Rieske iron-sulfur protein, validating the pathogenic effect of the UQCRFS1 variant. This report extends the genotypic and phenotypic spectrum for these two rare disorders and highlights the utility of deep phenotyping and genomics data to achieve diagnosis and insights into rare disease.
Keywords: GJA8; UQCRFS1; alopecia; cataracts; mitochondrial complex III; phenotypic spectrum; rare disease.
Figures



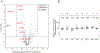
References
-
- Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, Xia F, Beaudet AL, Muzny DM, Gibbs RA, Boerwinkle E, Eng CM, Sutton VR, Shaw CA, Plon SE, Yang Y, Lupski JR, Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation, N. Engl. J. Med 376 (2017) 21–31, 10.1056/NEJMoa1516767. - DOI - PMC - PubMed
-
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM, Molecular findings among patients referred for clinical whole-exome sequencing, JAMA 312 (2014) 1870–1879, 10.1001/jama.2014.14601. - DOI - PMC - PubMed
-
- Balci TB, Hartley T, Xi Y, Dyment DA, Beaulieu CL, Bernier FP, Dupuis L, Horvath GA, Mendoza-Londono R, Prasad C, Richer J, Yang XR, Armour CM, Bareke E, Fernandez BA, McMillan HJ, Lamont RE, Majewski J, Parboosingh JS, Prasad AN, Rupar CA, Schwartzentruber J, Smith AC, Tetreault M, FORGE Canada Consortium, Care4Rare Canada Consortium, Innes AM, Boycott KM, Debunking Occam’s razor: Diagnosing multiple genetic diseases in families by whole-exome sequencing, Clin. Genet 92 (2017) 281–289, 10.1111/cge.12987. - DOI - PubMed
-
- Smith ED, Blanco K, Sajan SA, Hunter JM, Shinde DN, Wayburn B, Rossi M, Huang J, Stevens CA, Muss C, Alcaraz W, Hagman KDF, Tang S, Radtke K, A retrospective review of multiple findings in diagnostic exome sequencing, half are Distinct. half are overlapping Diagn.. Genet Med 21 (2019) 2199–2207, 10.1038/s41436-019-0477-2. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
