Stimulated Brillouin scattering flow cytometry
- PMID: 39421786
- PMCID: PMC11482170
- DOI: 10.1364/BOE.537602
Stimulated Brillouin scattering flow cytometry
Abstract
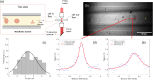
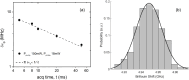
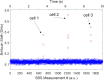
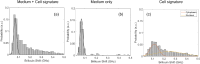
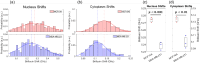
We present the use of stimulated Brillouin scattering spectroscopy to achieve rapid measurements of cell biomechanics in a flow cytometer setup. Specifically, our stimulated Brillouin scattering flow cytometry can acquire at a rate of 200 Hz, with a spectral acquisition time of 5 ms, which marks a 10x improvement compared to previous demonstrations of spontaneous Brillouin scattering flow cytometry. We experimentally validate our stimulated Brillouin scattering flow cytometer by measuring cell populations of normal breast epithelial cells and metastatic breast epithelial cancer cells.
© 2024 Optica Publishing Group.
Conflict of interest statement
None.
Figures
References
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
