Transcriptomic profiling of Schlemm's canal cells reveals a lymphatic-biased identity and three major cell states
- PMID: 39422453
- PMCID: PMC11488849
- DOI: 10.7554/eLife.96459
Transcriptomic profiling of Schlemm's canal cells reveals a lymphatic-biased identity and three major cell states
Abstract
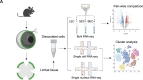
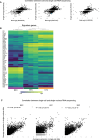
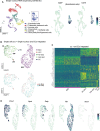
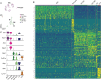
Schlemm's canal (SC) is central in intraocular pressure regulation but requires much characterization. It has distinct inner and outer walls, each composed of Schlemm's canal endothelial cells (SECs) with different morphologies and functions. Recent transcriptomic studies of the anterior segment added important knowledge, but were limited in power by SEC numbers or did not focus on SC. To gain a more comprehensive understanding of SC biology, we performed bulk RNA sequencing on C57BL/6 J SC, blood vessel, and lymphatic endothelial cells from limbal tissue (~4,500 SECs). We also analyzed mouse limbal tissues by single-cell and single-nucleus RNA sequencing (C57BL/6 J and 129/Sj strains), successfully sequencing 903 individual SECs. Together, these datasets confirm that SC has molecular characteristics of both blood and lymphatic endothelia with a lymphatic phenotype predominating. SECs are enriched in pathways that regulate cell-cell junction formation pointing to the importance of junctions in determining SC fluid permeability. Importantly, and for the first time, our analyses characterize three molecular classes of SECs, molecularly distinguishing inner wall from outer wall SECs and discovering two inner wall cell states that likely result from local environmental differences. Further, and based on ligand and receptor expression patterns, we document key interactions between SECs and cells of the adjacent trabecular meshwork (TM) drainage tissue. Also, we present cell type expression for a collection of human glaucoma genes. These data provide a new molecular foundation that will enable the functional dissection of key homeostatic processes mediated by SECs as well as the development of new glaucoma therapeutics.
Keywords: Schlemm's canal; cell biology; glaucoma anterior segment; mouse; single cell transcriptomics; trabecular meshwork.
© 2024, Balasubramanian, Kizhatil et al.
Conflict of interest statement
RB, KK, TL, NT, AB, GC, VB, RK, SZ, JP, MS, CM, WS, JQ, SJ No competing interests declared
Figures








Update of
-
Transcriptomic profiling of Schlemm's canal cells reveals a lymphatic-biased identity and three major cell states.bioRxiv [Preprint]. 2024 Aug 6:2023.08.31.555823. doi: 10.1101/2023.08.31.555823. bioRxiv. 2024. Update in: Elife. 2024 Oct 18;13:RP96459. doi: 10.7554/eLife.96459. PMID: 37886472 Free PMC article. Updated. Preprint.
References
-
- Aspelund A, Tammela T, Antila S, Nurmi H, Leppänen VM, Zarkada G, Stanczuk L, Francois M, Mäkinen T, Saharinen P, Immonen I, Alitalo K. The Schlemm’s canal is a VEGF-C/VEGFR-3-responsive lymphatic-like vessel. The Journal of Clinical Investigation. 2014;124:3975–3986. doi: 10.1172/JCI75395. - DOI - PMC - PubMed
-
- Barry DM, McMillan EA, Kunar B, Lis R, Zhang T, Lu T, Daniel E, Yokoyama M, Gomez-Salinero JM, Sureshbabu A, Cleaver O, Di Lorenzo A, Choi ME, Xiang J, Redmond D, Rabbany SY, Muthukumar T, Rafii S. Molecular determinants of nephron vascular specialization in the kidney. Nature Communications. 2019;10:5705. doi: 10.1038/s41467-019-12872-5. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
- CG2020004/BrightFocus Foundation
- P30 EY005722/EY/NEI NIH HHS/United States
- R01 EY032507/EY/NEI NIH HHS/United States
- R01EY022359/EY/NEI NIH HHS/United States
- R01EY029548/EY/NEI NIH HHS/United States
- P30 EY019007/EY/NEI NIH HHS/United States
- R01EY032507/EY/NEI NIH HHS/United States
- R01 EY022359/EY/NEI NIH HHS/United States
- R01EY028175/EY/NEI NIH HHS/United States
- R01 EY029548/EY/NEI NIH HHS/United States
- R01EY032062/EY/NEI NIH HHS/United States
- EMPIRE CU19-2660/New York Fund for Innovation in Research and Scientific Talent
- R01 EY032062/EY/NEI NIH HHS/United States
- G2021007S/BrightFocus Foundation
- R01 EY028175/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

