Re-emergence of Oropouche virus between 2023 and 2024 in Brazil: an observational epidemiological study
- PMID: 39423838
- PMCID: PMC11779697
- DOI: 10.1016/S1473-3099(24)00619-4
Re-emergence of Oropouche virus between 2023 and 2024 in Brazil: an observational epidemiological study
Abstract
Background: Oropouche virus is an arthropod-borne virus that has caused outbreaks of Oropouche fever in central and South America since the 1950s. This study investigates virological factors contributing to the re-emergence of Oropouche fever in Brazil between 2023 and 2024.
Methods: In this observational epidemiological study, we combined multiple data sources for Oropouche virus infections in Brazil and conducted in-vitro and in-vivo characterisation. We collected serum samples obtained in Manaus City, Amazonas state, Brazil, from patients with acute febrile illnesses aged 18 years or older who tested negative for malaria and samples from people with previous Oropouche virus infection from Coari municipality, Amazonas state, Brazil. Basic clinical and demographic data were collected from the Brazilian Laboratory Environment Management System. We calculated the incidence of Oropouche fever cases with data from the Brazilian Ministry of Health and the 2022 Brazilian population census and conducted age-sex analyses. We used reverse transcription quantitative PCR to test for Oropouche virus RNA in samples and subsequently performed sequencing and phylogenetic analysis of viral isolates. We compared the phenotype of the 2023-24 epidemic isolate (AM0088) with the historical prototype strain BeAn19991 through assessment of titre, plaque number, and plaque size. We used a plaque reduction neutralisation test (PRNT50) to assess the susceptibility of the novel isolate and BeAn19991 isolate to antibody neutralisation, both in serum samples from people previously infected with Oropouche virus and in blood collected from mice that were inoculated with either of the strains.
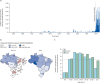
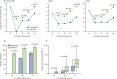
Findings: 8639 (81·8%) of 10 557 laboratory-confirmed Oropouche fever cases from Jan 4, 2015, to Aug 10, 2024, occurred in 2024, which is 58·8 times the annual median of 147 cases (IQR 73-325). Oropouche virus infections were reported in all 27 federal units, with 8182 (77·5%) of 10 557 infections occurring in North Brazil. We detected Oropouche virus RNA in ten (11%) of 93 patients with acute febrile illness between Jan 1 and Feb 4, 2024, in Amazonas state. AM0088 had a significantly higher replication at 12 h and 24 h after infection in mammalian cells than the prototype strain. AM0088 had a more virulent phenotype than the prototype in mammalian cells, characterised by earlier plaque formation, between 27% and 65% increase in plaque number, and plaques between 2·4-times and 2·6-times larger. Furthermore, serum collected on May 2 and May 20, 2016, from individuals previously infected with Oropouche virus showed at least a 32-fold reduction in neutralising capacity (ie, median PRNT50 titre of 640 [IQR 320-640] for BeAn19991 vs <20 [ie, below the limit of detection] for AM0088) against the reassortant strain compared with the prototype.
Interpretation: These findings provide a comprehensive assessment of Oropouche fever in Brazil and contribute to an improved understanding of the 2023-24 Oropouche virus re-emergence. Our exploratory in-vitro data suggest that the increased incidence might be related to a higher replication efficiency of a new Oropouche virus reassortant for which previous immunity shows lower neutralising capacity.
Funding: São Paulo Research Foundation, Burroughs Wellcome Fund, Wellcome Trust, US National Institutes of Health, and Brazilian National Council for Scientific and Technological Development.
Translation: For the Portuguese translation of the abstract see Supplementary Materials section.
Copyright © 2025 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures


Update of
-
Reemergence of Oropouche virus between 2023 and 2024 in Brazil.medRxiv [Preprint]. 2024 Jul 30:2024.07.27.24310296. doi: 10.1101/2024.07.27.24310296. medRxiv. 2024. Update in: Lancet Infect Dis. 2025 Feb;25(2):166-175. doi: 10.1016/S1473-3099(24)00619-4. PMID: 39132482 Free PMC article. Updated. Preprint.
References
-
- The Lancet Infectious Diseases Oropouche fever, the mysterious threat. Lancet Infect Dis. 2024;24:935. - PubMed
-
- Iani FCdM, Mota Pereira F, de Oliveira EC, et al. Rapid viral expansion beyond the Amazon Basin: increased epidemic activity of Oropouche virus across the Americas. medRxiv. 2024 doi: 10.1101/2024.08.02.24311415. published online Aug 6. (preprint). - DOI
-
- Pinheiro FP, Travassos da Rosa AP, Gomes ML, LeDuc JW, Hoch AL. Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science. 1982;215:1251–1253. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

