Novel, soluble 3-heteroaryl-substituted tanshinone mimics attenuate the inflammatory response in murine macrophages
- PMID: 39424621
- PMCID: PMC11489580
- DOI: 10.1038/s41598-024-73309-8
Novel, soluble 3-heteroaryl-substituted tanshinone mimics attenuate the inflammatory response in murine macrophages
Abstract
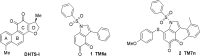
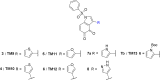
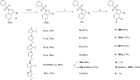
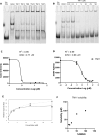
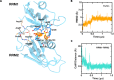
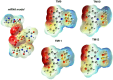
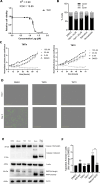
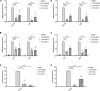
The RNA binding protein Human Antigen R (HuR) has been identified as a main regulator of the innate immune response and its inhibition can lead to beneficial anti-inflammatory effects. To this aim, we previously synthesized a novel class of small molecules named Tanshinone Mimics (TMs) able to interfere with HuR-RNA binding, and that dampen the LPS-induced immune response. Herein, we present a novel series of TMs, encompassing thiophene 3/TM9 and 4/TM10, furan 5/TM11 and 6/TM12, pyrrole 7b/TM13, and pyrazole 8. The furan-containing 5(TM11) showed the greatest inhibitory effect of the series on HuR-RNA complex formation, as suggested by RNA Electromobility Shift Assay and Time-Resolved FRET. Molecular Dynamics Calculation of HuR - 5/TM11 interaction, quantum mechanics approaches and Surface Plasmon Resonance data, all indicates that, within the novel heteroaryl substituents, the furan ring better recapitulates the chemical features of the RNA bound to HuR. Compound 5/TM11 also showed improved aqueous solubility compared to previously reported TMs. Real-time monitoring of cell growth and flow cytometry analyses showed that 5/TM11 preferentially reduced cell proliferation rather than apoptosis in murine macrophages at immunomodulatory doses. We observed its effects on the innate immune response triggered by lipopolysaccharide (LPS) in macrophages, showing that 5/TM11 significantly reduced the expression of proinflammatory cytokines as Cxcl10 and Il1b.
Keywords: Anti-inflammatory agents; ELAVL1; HuR; HuR inhibitors; LPS; Tanshinone mimics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
- GR2019-12369266/Ministero della Salute
- LM2023042/Ministerstvo Školství, Mládeže a Tělovýchovy
- . CZ.02.1.01/0.0/0.0/18_046/0015974/European Regional Development Fund
- PNRR-MUR NextGenerationEU cod. CN00000041/Ministero dell'Università e della Ricerca
- 21548/Associazione Italiana per la Ricerca sul Cancro
LinkOut - more resources
Full Text Sources
Miscellaneous

