TDP43 aggregation at ER-exit sites impairs ER-to-Golgi transport
- PMID: 39424779
- PMCID: PMC11489672
- DOI: 10.1038/s41467-024-52706-7
TDP43 aggregation at ER-exit sites impairs ER-to-Golgi transport
Abstract
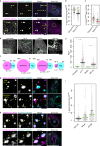
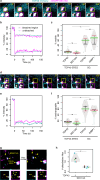
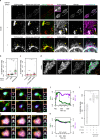
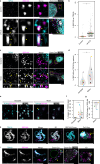
Protein aggregation plays key roles in age-related degenerative diseases, but how different proteins coalesce to form inclusions that vary in composition, morphology, molecular dynamics and confer physiological consequences is poorly understood. Here we employ a general reporter based on mutant Hsp104 to identify proteins forming aggregates in human cells under common proteotoxic stress. We identify over 300 proteins that form different inclusions containing subsets of aggregating proteins. In particular, TDP43, implicated in Amyotrophic Lateral Sclerosis (ALS), partitions dynamically between two distinct types of aggregates: stress granule and a previously unknown non-dynamic (solid-like) inclusion at the ER exit sites (ERES). TDP43-ERES co-aggregation is induced by diverse proteotoxic stresses and observed in the motor neurons of ALS patients. Such aggregation causes retention of secretory cargos at ERES and therefore delays ER-to-Golgi transport, providing a link between TDP43 aggregation and compromised cellular function in ALS patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Zeskind, B. J. et al. Nucleic acid and protein mass mapping by live-cell deep-ultraviolet microscopy. Nat. Methods4, 567–569 (2007). - PubMed
-
- Ellis, R. J. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol.11, 114–119 (2001). - PubMed
-
- Uversky, V. N. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol.44, 18–30 (2017). - PubMed
-
- Ellis, R. J. & Minton, A. P. Protein aggregation in crowded environments. Biol. Chem.387, 485–497 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- A-0007081-00-00/Ministry of Education - Singapore (MOE)
- NRF-MSG-2023-0001/National Research Foundation Singapore (National Research Foundation-Prime Minister's office, Republic of Singapore)
- NRF-SIS "SingMass"/A*STAR | Singapore Institute of Manufacturing Technology (Singapore Institute of Manufacturing Technology - A STAR)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

