Modal-nexus auto-encoder for multi-modality cellular data integration and imputation
- PMID: 39424861
- PMCID: PMC11489673
- DOI: 10.1038/s41467-024-53355-6
Modal-nexus auto-encoder for multi-modality cellular data integration and imputation
Abstract
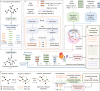
Heterogeneous feature spaces and technical noise hinder the cellular data integration and imputation. The high cost of obtaining matched data across modalities further restricts analysis. Thus, there's a critical need for deep learning approaches to effectively integrate and impute unpaired multi-modality single-cell data, enabling deeper insights into cellular behaviors. To address these issues, we introduce the Modal-Nexus Auto-Encoder (Monae). Leveraging regulatory relationships between modalities and employing contrastive learning within modality-specific auto-encoders, Monae enhances cell representations in the unified space. The integration capability of Monae furnishes it with modality-complementary cellular representations, enabling the generation of precise intra-modal and cross-modal imputation counts for extensive and complex downstream tasks. In addition, we develop Monae-E (Monae-Extension), a variant of Monae that can converge rapidly and support biological discoveries. Evaluations on various datasets have validated Monae and Monae-E's accuracy and robustness in multi-modality cellular data integration and imputation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Lv, Q., Chen, G., Yang, Z., Zhong, W., Chen, C.Y.-C. Meta learning with graph attention networks for low-data drug discovery. IEEE Trans. Neural Netw. Learn. Syst.35, 11218–11230 (2023). - PubMed
-
- Tang, Z., Chen, G., Yang, H., Zhong, W., Chen, C.Y.-C. DSIL-DDI: A domain-invariant substructure interaction learning for generalizable drug–drug interaction prediction. IEEE Trans. Neural Netw. Learn. Syst.35, 10552–10560 (2023). - PubMed
-
- Luecken, M.D. et al. A sandbox for prediction and integration of dna, rna, and proteins in single cells. In Thirty-fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 2) (2021).
-
- Patruno, L. et al. A review of computational strategies for denoising and imputation of single-cell transcriptomic data. Brief. Bioinforma.22, 222 (2021). - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

