The alternative splicing landscape of infarcted mouse heart identifies isoform level therapeutic targets
- PMID: 39424867
- PMCID: PMC11489681
- DOI: 10.1038/s41597-024-03998-3
The alternative splicing landscape of infarcted mouse heart identifies isoform level therapeutic targets
Abstract
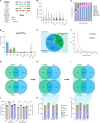
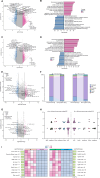
Alternative splicing is an important process that contributes to highly diverse transcripts and protein products, which can affect the development of disease in various organisms. Cardiovascular disease (CVD) represents one of the greatest global threats to humans, particularly acute myocardial infarction (MI) and subsequent ischemic reperfusion (IR) injury, which involve complex transcriptomic changes in heart tissues associated with metabolic reshaping and immunological response. In this study, we used a newly developed ONT full-length transcriptomic approach and performed transcript-resolved differential expression profiling in murine models of MI and IR. We built an analytical pipeline to reliably identify and quantify alternative splicing products (isoforms), expanding on the currently available catalog of isoforms described in mice. The updated alternative splicing landscape included transcripts, genes, and pathways that were differentially regulated during IR and MI. Our study establishes a pipeline to profile highly diverse isoforms using state-of-the-art long-read sequencing, builds a landscape of alternative splicing in the mouse heart during MI and IR.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Mechanic, O. J., Gavin, M. & Grossman, S. A. Acute Myocardial Infarction. in StatPearls (StatPearls Publishing, Treasure Island (FL), 2024). - PubMed
-
- Stähli, B. E. et al. Timing of Complete Revascularization with Multivessel PCI for Myocardial Infarction. N Engl J Med389, 1368–1379 (2023). - PubMed
-
- Barrère-Lemaire, S. et al. Mesenchymal stromal cells for improvement of cardiac function following acute myocardial infarction: a matter of timing. Physiol Rev104, 659–725 (2024). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

