Empirical assessment of the enrichment-based metagenomic methods in identifying diverse respiratory pathogens
- PMID: 39424897
- PMCID: PMC11489750
- DOI: 10.1038/s41598-024-75120-x
Empirical assessment of the enrichment-based metagenomic methods in identifying diverse respiratory pathogens
Abstract
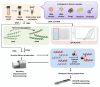
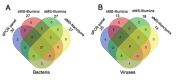
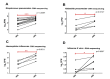
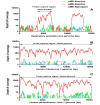
Probe-based nucleic acid enrichment represents an effective route to enhance the detection capacity of next-generation sequencing (NGS) in a set of clinically diverse and relevant microbial species. In this study, we assessed the effect of the enrichment-based sequencing on identifying respiratory infections using tiling RNA probes targeting 76 respiratory pathogens and sequenced using both Illumina and Oxford Nanopore platforms. Forty respiratory swab samples pre-tested for a panel of respiratory pathogens by qPCR were used to benchmark the sequencing data. We observed a general improvement in sensitivity after enrichment. The overall detection rate increased from 73 to 85% after probe capture detected by Illumina. Moreover, enrichment with probe sets boosted the frequency of unique pathogen reads by 34.6 and 37.8-fold for Illumina DNA and cDNA sequencing, respectively. This also resulted in significant improvements on genome coverage especially in viruses. Despite these advantages, we found that library pooling may cause reads mis-assignment, probably due to crosstalk issues arise from post-capture PCR and from pooled sequencing, thus increasing the risk of bleed-through signal. Taken together, an overall improvement in the breadth and depth of pathogen coverage is achieved using enrichment-based sequencing method. For future applications, automated library processing and pooling-free sequencing could enhance the precision and timeliness of probe enrichment-based clinical metagenomics.
Keywords: Illumina; Nanopore; Respiratory pathogens; Virus; mNGS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Metagenomic next-generation sequencing targeted and metagenomic next-generation sequencing for pulmonary infection in HIV-infected and non-HIV-infected individuals.Front Cell Infect Microbiol. 2024 Aug 19;14:1438982. doi: 10.3389/fcimb.2024.1438982. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39224706 Free PMC article.
-
Targeted Sequencing of Respiratory Viruses in Clinical Specimens for Pathogen Identification and Genome-Wide Analysis.Methods Mol Biol. 2018;1838:125-140. doi: 10.1007/978-1-4939-8682-8_10. Methods Mol Biol. 2018. PMID: 30128994 Free PMC article.
-
Comparative analysis of metagenomic and targeted next-generation sequencing for pathogens diagnosis in bronchoalveolar lavage fluid specimens.Front Cell Infect Microbiol. 2024 Aug 27;14:1451440. doi: 10.3389/fcimb.2024.1451440. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39258254 Free PMC article.
-
Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections.J Adv Res. 2021 Sep 29;38:201-212. doi: 10.1016/j.jare.2021.09.012. eCollection 2022 May. J Adv Res. 2021. PMID: 35572406 Free PMC article. Review.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
References
-
- Mitchell, A. B. & Glanville, A. R. Introduction to techniques and methodologies for characterizing the human respiratory virome. Methods Mol. Biol.1838, 111–123 (2018). - PubMed
MeSH terms
Grants and funding
- 81801991/the National Natural Science Foundation of China
- GW VI-11.1-09/three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai
- GWVI-2.2/three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai
- GWVI-11.1-15/three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai
- 20Y11911300/the 2020 Shanghai Science and Technology Innovation Action Plan Medical Innovation Research Special Project
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

