A SIRT7-dependent acetylation switch regulates early B cell differentiation and lineage commitment through Pax5
- PMID: 39424985
- PMCID: PMC11588656
- DOI: 10.1038/s41590-024-01995-7
A SIRT7-dependent acetylation switch regulates early B cell differentiation and lineage commitment through Pax5
Abstract
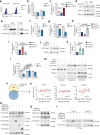
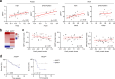
B lymphopoiesis is orchestrated by lineage-specific transcription factors. In B cell progenitors, lineage commitment is mediated by Pax5, which is commonly mutated in B cell acute lymphoblastic leukemia. Despite its essential role in immunity, the mechanisms regulating Pax5 function remain largely unknown. Here, we found that the NAD+-dependent enzyme SIRT7 coordinates B cell development through deacetylation of Pax5 at K198, which promotes Pax5 protein stability and transcriptional activity. Neither Pax5K198 deacetylated nor acetylated mimics rescued B cell differentiation in Pax5-/- pro-B cells, suggesting that B cell development requires Pax5 dynamic deacetylation. The Pax5K198 deacetylation mimic restored lineage commitment in Pax5-/- pro-B cells and B cell differentiation in Sirt7-/- pro-B cells, suggesting the uncoupling of differentiation from lineage commitment. The SIRT7-Pax5 interplay was conserved in B cell acute lymphoblastic leukemia, where SIRT7 expression correlated with good prognosis. Our findings reveal a crucial mechanism for B lymphopoiesis and highlight the relevance of sirtuins in immune function.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




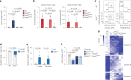

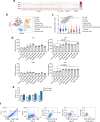
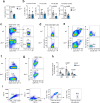

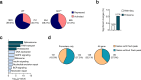


References
-
- Medvedovic, J., Ebert, A., Tagoh, H. & Busslinger, M. PAX5: a master regulator of B cell development and leukemogenesis. Adv. Immunol.111, 179–206 (2011). - PubMed
-
- Delogu, A. et al. Gene repression by PAX5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity24, 269–281 (2006). - PubMed
-
- Cobaleda, C., Schebesta, A., Delogu, A. & Busslinger, M. PAX5: the guardian of B cell identity and function. Nat. Immunol.8, 463–470 (2007). - PubMed
-
- Schebesta, A. et al. Transcription factor PAX5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity27, 49–63 (2007). - PubMed
-
- Urbanek, P., Wang, Z. Q., Fetka, I., Wagner, E. F. & Busslinger, M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking PAX5/BSAP. Cell79, 901–912 (1994). - PubMed
MeSH terms
Substances
Grants and funding
- PROYE20042VAQU/Fundación Científica Asociación Española Contra el Cáncer (Scientific Foundation, Spanish Association Against Cancer)
- SAF2017-88975R/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- 2017-SGR-148/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- 2021-SGR-01378/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- FI_B 00293/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- 2021-SGR-01494/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- 2016-BP-00250/Departament d'Innovació, Universitats i Empresa, Generalitat de Catalunya (Department of Innovation, Education and Enterprise, Government of Catalonia)
- MSC-2021-101065013/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- MSC-2020-895979/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- 2022-0019/Barncancerfonden (Swedish Childhood Cancer Foundation)
- 2021-02379/Forskningsrådet om Hälsa, Arbetsliv och Välfärd (Swedish Research Council for Health, Working Life and Welfare)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

