New perspectives on YTHDF2 O-GlcNAc modification in the pathogenesis of intervertebral disc degeneration
- PMID: 39425013
- PMCID: PMC11488288
- DOI: 10.1186/s10020-024-00876-x
New perspectives on YTHDF2 O-GlcNAc modification in the pathogenesis of intervertebral disc degeneration
Abstract
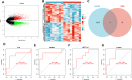
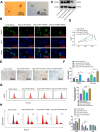
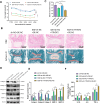
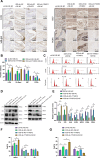
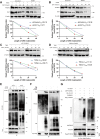
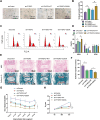
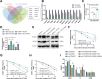
This study investigates the potential molecular mechanisms by which O-GlcNAc modification of YTHDF2 regulates the cell cycle and participates in intervertebral disc degeneration (IDD). We employed transcriptome sequencing to identify genes involved in IDD and utilized bioinformatics analysis to predict key disease-related genes. In vitro mechanistic validation was performed using mouse nucleus pulposus (NP) cells. Changes in reactive oxygen species (ROS) and cell cycle were assessed through flow cytometry and CCK-8 assays. An IDD mouse model was also established for in vivo mechanistic validation, with changes in IDD severity measured using X-rays and immunohistochemical staining. Bioinformatics analysis revealed differential expression of YTHDF2 in NP cells of normal and IDD mice, suggesting its potential as a diagnostic gene for IDD. In vitro cell experiments demonstrated that YTHDF2 expression and O-GlcNAcylation were reduced in NP cells under H2O2 induction, leading to inhibition of the cell cycle through decreased stability of CCNE1 mRNA. Further, in vivo animal experiments confirmed a decrease in YTHDF2 expression and O-GlcNAcylation in IDD mice, while overexpression or increased O-GlcNAcylation of YTHDF2 promoted CCNE1 protein expression, thereby alleviating IDD pathology. YTHDF2 inhibits its degradation through O-GlcNAc modification, promoting the stability of CCNE1 mRNA and the cell cycle to prevent IDD formation.
Keywords: CCNE1; Cell cycle; Intervertebral disc degeneration; O-GlcNAc modification; Oxidative stress; YTHDF2.
© 2024. The Author(s).
Conflict of interest statement
The author declares no conflict of interest.
Figures




References
-
- Cao S, Liu H, Fan J, et al. An oxidative stress-related gene pair (CCNB1/PKD1), competitive endogenous RNAs, and Immune-infiltration patterns potentially regulate intervertebral disc Degeneration Development. Front Immunol. 2021;12:765382. 10.3389/fimmu.2021.765382. Published 2021 Nov 9. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

