COL25A1 and METAP1D DNA methylation are promising liquid biopsy epigenetic biomarkers of colorectal cancer using digital PCR
- PMID: 39425144
- PMCID: PMC11490026
- DOI: 10.1186/s13148-024-01748-1
COL25A1 and METAP1D DNA methylation are promising liquid biopsy epigenetic biomarkers of colorectal cancer using digital PCR
Abstract
Background: Colorectal cancer is a public health issue and was the third leading cause of cancer-related death worldwide in 2022. Early diagnosis can improve prognosis, making screening a central part of colorectal cancer management. Blood-based screening, diagnosis and follow-up of colorectal cancer patients are possible with the study of cell-free circulating tumor DNA. This study aimed to identify novel DNA methylation biomarkers of colorectal cancer that can be used for the follow-up of patients with colorectal cancer.
Methods: A DNA methylation profile was established in the Gene Expression Omnibus (GEO) database (n = 507) using bioinformatics analysis and subsequently confirmed using The Cancer Genome Atlas (TCGA) database (n = 348). The in silico profile was then validated on local tissue and cell-free DNA samples using methylation-specific digital PCR in colorectal cancer patients (n = 35) and healthy donors (n = 35).
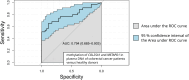
Results: The DNA methylation of COL25A1 and METAP1D was predicted to be a colorectal cancer biomarker by bioinformatics analysis (ROC AUC = 1, 95% CI [0.999-1]). The two biomarkers were confirmed with tissue samples, and the combination of COL25A1 and METAP1D yielded 49% sensitivity and 100% specificity for cell-free DNA.
Conclusion: Bioinformatics analysis of public databases revealed COL25A1 and METAP1D DNA methylation as clinically applicable liquid biopsies DNA methylation biomarkers. The specificity implies an excellent positive predictive value for follow-up, and the high sensitivity and relative noninvasiveness of a blood-based test make these biomarkers compatible with colorectal cancer screening. However, the clinical impact of these biomarkers in colorectal cancer screening and follow-up needs to be established in further prospective studies.
Keywords: Biomarker; COL25A1; Circulating tumor DNA; Colorectal cancer; DNA methylation; Digital PCR; Liquid biopsy; METAP1D.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Ferlay J, Ervik M, Lam F, Laversanne M, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F. Global cancer observatory: cancer today. Lyon: International Agency for Research on Cancer; 2024. Available from: https://gco.iarc.who.int/today, Accessed 06 March 2024.
-
- Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. - PubMed
-
- Winawer SJ, Zauber AG. The advanced adenoma as the primary target of screening. Gastrointest Endosc Clin N Am. 2002;12(1–9):v. - PubMed
-
- Tinmouth J, Lansdorp-Vogelaar I, Allison JE. Faecal immunochemical tests versus guaiac faecal occult blood tests: what clinicians and colorectal cancer screening programme organisers need to know. Gut. 2015;64:1327–37. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

