JunB is required for CD8+ T cell responses to acute infections
- PMID: 39425978
- PMCID: PMC11884676
- DOI: 10.1093/intimm/dxae063
JunB is required for CD8+ T cell responses to acute infections
Abstract
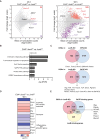
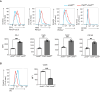
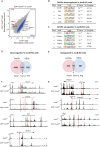
Basic-leucine zipper transcription factor ATF-like (BATF) and interferon regulatory factor 4 (IRF4) are crucial transcription factors for the generation of cytotoxic effector and memory CD8+ T cells. JunB is required for expression of genes controlled by BATF and IRF4 in CD4+ T cell responses, but the role of JunB in CD8+ T cells remains unknown. Here, we demonstrate that JunB is essential for cytotoxic CD8+ T cell responses. JunB expression is transiently induced, depending on the T cell receptor signal strength. JunB deficiency severely impairs the clonal expansion of effector CD8+ T cells in response to acute infection with Listeria monocytogenes. Junb-deficient CD8+ T cells fail to control transcription and chromatin accessibility of a specific set of genes regulated by BATF and IRF4, resulting in impaired cell survival, glycolysis, and cytotoxic CD8+ T cell differentiation. Furthermore, JunB deficiency enhances the expression of co-inhibitory receptors, including programmed cell death 1 (PD-1) and T cell immunoglobulin mucin-3 (TIM3) upon activation of naive CD8+ T cells. These results indicate that JunB, in collaboration with BATF and IRF4, promotes multiple key events in the early stage of cytotoxic CD8+ T cell responses.
Keywords: AP-1; apoptosis; co-inhibitory molecules; effector and memory CD8+ T cells; glycolysis.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Japanese Society for Immunology.
Figures




References
-
- Reina-Campos M, Scharping NE, Goldrath AW.. CD8(+) T cell metabolism in infection and cancer. Nat Rev Immunol 2021;21:718–38. https://doi.org/10.1038/s41577-021-00537-8 - DOI - PMC - PubMed
-
- Williams MA, Bevan MJ.. Effector and memory CTL differentiation. Annu Rev Immunol 2007;25:171–92. https://doi.org/10.1146/annurev.immunol.25.022106.141548 - DOI - PubMed
-
- Cui W, Kaech SM.. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev 2010;236:151–66. https://doi.org/10.1111/j.1600-065X.2010.00926.x - DOI - PMC - PubMed
-
- Kaech SM, Wherry EJ.. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 2007;27:393–405. https://doi.org/10.1016/j.immuni.2007.08.007 - DOI - PMC - PubMed
-
- Joshi NS, Cui W, Chandele A, et al.Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 2007;27:281–95. https://doi.org/10.1016/j.immuni.2007.07.010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

