In vitro analysis suggests that SARS-CoV-2 infection differentially modulates cancer-like phenotypes and cytokine expression in colorectal and prostate cancer cells
- PMID: 39427065
- PMCID: PMC11490510
- DOI: 10.1038/s41598-024-75718-1
In vitro analysis suggests that SARS-CoV-2 infection differentially modulates cancer-like phenotypes and cytokine expression in colorectal and prostate cancer cells
Abstract
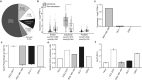
The coronavirus disease 2019 (COVID-19) reportedly exacerbates cancer outcomes. However, how COVID-19 influences cancer prognosis and development remains poorly understood. Here, we investigated the effect of Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2), the etiological agent of COVID-19, on cellular cancer phenotypes the expression of cancer-related markers, and various proinflammatory cytokines. We infected prostate (22RV1) and colorectal (DLD-1) cancer cell lines, which express angiotensin-converting enzyme 2 (ACE2), with spike pseudovirus (sPV) and laboratory stocks of live SARS-CoV-2 viruses. After infection, we quantified changes in the cellular cancer phenotypes, the gene expression levels of some cancer markers, including Ki-67, BCL-2, VIM, MMP9, and VEGF, and proinflammatory cytokines. Phenotypic analysis was performed using MTT and wound healing assays, whereas gene expression analysis was carried out using real-time quantitative PCR (RT-qPCR). We show that SARS-CoV-2 infection impacts several key cellular pathways involved in cell growth, apoptosis, and migration, in prostate and colorectal cancer cells. Our results suggest that SARS-CoV-2 infection does influence various cancer cellular phenotypes and expression of molecular cancer markers and proinflammatory cytokines, albeit in a cell-type-specific manner. Our findings hint at the need for further studies and could have implications for evaluating the impact of other viruses on cancer progression.
Keywords: COVID-19; Cancer; Cytokine expression; SARS-CoV-2; Spike pseudovirus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


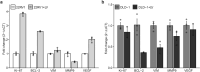

References
-
- Bennett, J. E. et al. NCD Countdown 2030: Worldwide trends in non-communicable disease mortality and progress towards Sustainable Development Goal target 3.4. Lancet392, 1072–1088 (2018). - PubMed
-
- O’Donnell, J. S., Teng, M. W. L. & Smyth, M. J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol.16, 151–167 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

