Transmission of highly virulent CXCR4 tropic HIV-1 through the mucosal route in an individual with a wild-type CCR5 genotype
- PMID: 39427414
- PMCID: PMC11533037
- DOI: 10.1016/j.ebiom.2024.105410
Transmission of highly virulent CXCR4 tropic HIV-1 through the mucosal route in an individual with a wild-type CCR5 genotype
Abstract
Background: Nearly all transmitted/founder (T/F) HIV-1 are CCR5 (R5)-tropic. While previous evidence suggested that CXCR4 (X4)-tropic HIV-1 are transmissible, virus detection and characterization were not at the earliest stages of acute infection.
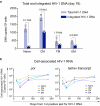
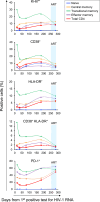
Methods: We identified an X4-tropic T/F HIV-1 in a participant (40700) in the RV217 acute infection cohort. Coreceptor usage was determined in TZM-bl cell line, NP-2 cell lines, and primary CD4+ T cells using pseudovirus and infectious molecular clones. CD4 subset dynamics were analyzed using flow cytometry. Viral load in each CD4 subset was quantified using cell-associated HIV RNA assay and total and integrated HIV DNA assay.
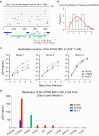
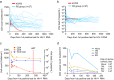
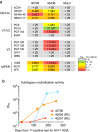
Findings: Participant 40700 was infected by an X4 tropic HIV-1 without CCR5 using ability. This participant experienced significantly faster CD4 depletion compared to R5 virus infected individuals in the same cohort. Naïve and central memory (CM) CD4 subsets declined faster than effector memory (EM) and transitional memory (TM) subsets. All CD4 subsets, including the naïve, were productively infected. Increased CD4+ T cell activation was observed over time. This X4-tropic T/F virus is resistant to broadly neutralizing antibodies (bNAbs) targeting V1/V2 and V3 regions, while most of the R5 T/F viruses in the same cohort are sensitive to the same panel of bNAbs.
Interpretation: X4-tropic HIV-1 is transmissible through mucosal route in people with wild-type CCR5 genotype. The CD4 subset tropism of HIV-1 may be an important determinant for HIV-1 transmissibility and virulence.
Funding: Institute of Human Virology, National Institutes of Health, Henry M. Jackson Foundation for the Advancement of Military Medicine.
Keywords: CD4 subset; CXCR4; HIV-1; Mucosal transmission; Pathogenesis.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Update of
-
Transmission of highly virulent CXCR4 tropic HIV-1 through the mucosal route in an individual with a wild-type CCR5 genotype.bioRxiv [Preprint]. 2023 Sep 15:2023.09.15.557832. doi: 10.1101/2023.09.15.557832. bioRxiv. 2023. Update in: EBioMedicine. 2024 Nov;109:105410. doi: 10.1016/j.ebiom.2024.105410. PMID: 37745406 Free PMC article. Updated. Preprint.
-
Transmission of highly virulent CXCR4 tropic HIV-1 through the mucosal route in an individual with a wild-type CCR5 genotype.Res Sq [Preprint]. 2023 Sep 25:rs.3.rs-3359209. doi: 10.21203/rs.3.rs-3359209/v1. Res Sq. 2023. Update in: EBioMedicine. 2024 Nov;109:105410. doi: 10.1016/j.ebiom.2024.105410. PMID: 37841838 Free PMC article. Updated. Preprint.
References
-
- Smolen-Dzirba J., Rosinska M., Janiec J., et al. HIV-1 infection in persons homozygous for CCR5-delta 32 allele: the next case and the review. AIDS Rev. 2017;19(4):219–230. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

