Chromosomal reference genome sequences for the malaria mosquito, Anopheles coustani, Laveran, 1900
- PMID: 39429628
- PMCID: PMC11490835
- DOI: 10.12688/wellcomeopenres.22983.1
Chromosomal reference genome sequences for the malaria mosquito, Anopheles coustani, Laveran, 1900
Abstract
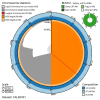
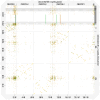
We present genome assembly from individual female An. coustani (African malaria mosquito; Arthropoda; Insecta; Diptera; Culicidae) from Lopé, Gabon. The genome sequence is 270 megabases in span. Most of the assembly is scaffolded into three chromosomal pseudomolecules with the X sex chromosome assembled for both species. The complete mitochondrial genome was also assembled and is 15.4 kilobases in length.
Keywords: African malaria mosquito; Anopheles coustani; chromosomal; genome sequence.
Copyright: © 2024 Bouafou LBA et al.
Conflict of interest statement
No competing interests were disclosed.
Figures



References
-
- Gillies MT, Coetzee M: A supplement to the anophelinae of Africa south of the Sahara (Afrotropical region).THE SOUTH AFRICAN INSTITUTE FOR MEDICAL RESEARCH,1987. Reference Source - DOI
-
- Laveran A: Sur un anopheles provenant de Madagascar. C R Seances Soc Biol Fil. 1900;57:109–110.
Grants and funding
LinkOut - more resources
Full Text Sources

