Surpassing the Diffraction Limit in Label-Free Optical Microscopy
- PMID: 39429866
- PMCID: PMC11487630
- DOI: 10.1021/acsphotonics.4c00745
Surpassing the Diffraction Limit in Label-Free Optical Microscopy
Abstract
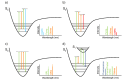
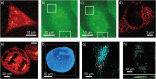
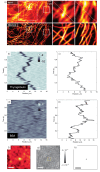
Super-resolution optical microscopy has enhanced our ability to visualize biological structures on the nanoscale. Fluorescence-based techniques are today irreplaceable in exploring the structure and dynamics of biological matter with high specificity and resolution. However, the fluorescence labeling concept narrows the range of observed interactions and fundamentally limits the spatiotemporal resolution. In contrast, emerging label-free imaging methods are not inherently limited by speed and have the potential to capture the entirety of complex biological processes and dynamics. While pushing a complex unlabeled microscopy image beyond the diffraction limit to single-molecule resolution and capturing dynamic processes at biomolecular time scales is widely regarded as unachievable, recent experimental strides suggest that elements of this vision might be already in place. These techniques derive signals directly from the sample using inherent optical phenomena, such as elastic and inelastic scattering, thereby enabling the measurement of additional properties, such as molecular mass, orientation, or chemical composition. This perspective aims to identify the cornerstones of future label-free super-resolution imaging techniques, discuss their practical applications and theoretical challenges, and explore directions that promise to enhance our understanding of complex biological systems through innovative optical advancements. Drawing on both traditional and emerging techniques, label-free super-resolution microscopy is evolving to offer detailed and dynamic imaging of living cells, surpassing the capabilities of conventional methods for visualizing biological complexities without the use of labels.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
