NMR metabolomics of plant and yeast-based hydrolysates for cell culture media applications - A comprehensive assessment
- PMID: 39429919
- PMCID: PMC11490674
- DOI: 10.1016/j.crfs.2024.100855
NMR metabolomics of plant and yeast-based hydrolysates for cell culture media applications - A comprehensive assessment
Abstract
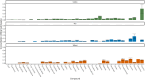
Cultivated meat products, generated by growing isolated skeletal muscle and fat tissue, offer the promise of a more sustainable and ethical alternative to traditional meat production. However, with cell culture media used to grow the cells accounting for 55-95% of the overall production cost, achieving true sustainability requires significant media optimization. One means of dealing with these high costs is the use of low-cost complex additives such as hydrolysates to provide a wide range of nutrients, from small molecules (metabolites) to growth factors and peptides. Despite their potential, most hydrolysate products remain poorly characterized and many are thought to suffer from persistent issues of high batch-to-batch variability. Although there have been a number of isolated efforts to determine metabolic profiles for a handful of hydrolysate products, we present the first attempt at a more comprehensive metabolomic characterization of nine different products (four plant and five yeast-based) from two to four different lots each. NMR analysis identified 90 unique metabolites, with only 15 metabolites common to all hydrolysate products (including eight of the nine essential amino acids), and 16 metabolites found in only a single hydrolysate product. The different hydrolysate products were found to have substantial differences in metabolite concentrations (as a fraction of overall mass), ranging from a high of 43% in yeast extract to a low of 14% in soy hydrolysates. The proportion of various metabolites also varied between products, with carbohydrate concentrations particularly high in soy hydrolysates and nucleosides more prominent in two of the yeast products. Overall, yeast extract generally had higher metabolite concentrations than all the other products, whereas both yeast extract and cotton had the largest variety of metabolites. A direct calculation of batch-to-batch variability revealed although there are significant differences between lots, these are largely driven by a relatively small fraction of compounds. This report will hopefully serve as a useful starting point for a more nuanced consideration of hydrolysate products in cell culture media optimization, both in the context of cultivated meat and beyond.
Keywords: Cultivated meat; Hydrolysates; Metabolomics; Nuclear magnetic resonance.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Stanislav Sokolenko reports equipment, drugs, or supplies was provided by Kerry Group. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Albertyn J., Hohmann S., Prior B.A. Characterization of the osmotic-stress response in saccharomyces cerevisiae: osmotic stress and glucose repression regulate glycerol-3-phosphate dehydrogenase independently. Curr. Genet. 1994;25:12–18. 1. - PubMed
-
- Bahieldin A., Sabir J.S., Ramadan A., Alzohairy A.M., Younis R.A., Shokry A.M., Gadalla N.O., Edris S., Hassan S.M., Al-Kordy M.A., Kamal K.B., Rabah S., Abuzinadah O.A., El-Domyati F.M. Control of glycerol biosynthesis under high salt stress in Arabidopsis. Funct. Plant Biol. 2014;41:87–95. - PubMed
-
- Djemal L., von Hagen J., Kolmar H., Deparis V. Characterization of soy protein hydrolysates and influence of its iron content on monoclonal antibody production by a murine hybridoma cell line. Biotechnol. Prog. 2021;37 7. - PubMed
-
- Gilbert A., Huang Y.-M., Ryll T. Identifying and eliminating cell culture process variability. Pharmaceut. Bioprocess. 2014;2:519–534.
LinkOut - more resources
Full Text Sources

