Advancing Anticancer Drug Discovery: Leveraging Metabolomics and Machine Learning for Mode of Action Prediction by Pattern Recognition
- PMID: 39431333
- PMCID: PMC11653622
- DOI: 10.1002/advs.202404085
Advancing Anticancer Drug Discovery: Leveraging Metabolomics and Machine Learning for Mode of Action Prediction by Pattern Recognition
Abstract
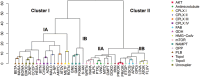
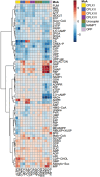
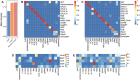
A bottleneck in the development of new anti-cancer drugs is the recognition of their mode of action (MoA). Metabolomics combined with machine learning allowed to predict MoAs of novel anti-proliferative drug candidates, focusing on human prostate cancer cells (PC-3). As proof of concept, 38 drugs are studied with known effects on 16 key processes of cancer metabolism, profiling low molecular weight intermediates of the central carbon and cellular energy metabolism (CCEM) by LC-MS/MS. These metabolic patterns unveiled distinct MoAs, enabling accurate MoA predictions for novel agents by machine learning. The transferability of MoA predictions based on PC-3 cell treatments is validated with two other cancer cell models, i.e., breast cancer and Ewing's sarcoma, and show that correct MoA predictions for alternative cancer cells are possible, but still at some expense of prediction quality. Furthermore, metabolic profiles of treated cells yield insights into intracellular processes, exemplified for drugs inducing different types of mitochondrial dysfunction. Specifically, it is predicted that pentacyclic triterpenes inhibit oxidative phosphorylation and affect phospholipid biosynthesis, as confirmed by respiration parameters, lipidomics, and molecular docking. Using biochemical insights from individual drug treatments, this approach offers new opportunities, including the optimization of combinatorial drug applications.
Keywords: cancer; drug discovery; machine learning; metabolomics; mode of action.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
