Phenotypic diversity of type III secretion system activity in enteropathogenic Escherichia coli clinical isolates
- PMID: 39432330
- PMCID: PMC11493143
- DOI: 10.1099/jmm.0.001907
Phenotypic diversity of type III secretion system activity in enteropathogenic Escherichia coli clinical isolates
Abstract
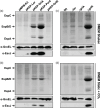
Introduction. Enteropathogenic Escherichia coli (EPEC) strains pose a significant threat as a leading cause of severe childhood diarrhoea in developing nations. EPEC pathogenicity relies on the type III secretion system (T3SS) encoded by the locus of enterocyte effacement (LEE), facilitating the secretion and translocation of bacterial effector proteins.Gap Statement. While the regulatory roles of PerC (plasmid-encoded regulator) and GrlA (global regulator of LEE-activator) in ler expression and LEE gene activation are well-documented in the EPEC prototype strain E2348/69, understanding the variability in LEE gene expression control mechanisms among clinical EPEC isolates remains an area requiring further investigation.Aim. This study aims to explore the diversity in LEE gene expression control mechanisms among clinical EPEC isolates through a comparative analysis of secretion profiles under defined growth conditions favouring either PerC- or GrlA-mediated activation of LEE expression.Methodology. We compared T3SS-dependent secretion patterns and promoter expression in both typical EPEC (tEPEC) and atypical EPEC (aEPEC) clinical isolates under growth conditions favouring either PerC- or GrlA-mediated activation of LEE expression. Additionally, we conducted promoter reporter activity assays, quantitative real-time PCR and Western blot experiments to assess gene expression activity.Results. Significant differences in T3SS-dependent secretion were observed among tEPEC and aEPEC strains, independent of LEE sequence variations or T3SS gene functionality. Notably, a clinical tEPEC isolate exhibited increased secretion levels under repressive growth conditions and in the absence of both PerC and GrlA, implicating an alternative mechanism in the activation of Ler (LEE-encoded regulator) expression.Conclusion. Our findings indicate that uncharacterized LEE regulatory mechanisms contribute to phenotypic diversity among clinical EPEC isolates, though their impact on clinical outcomes remains unknown. This challenges the conventional understanding based on reference strains and highlights the need to investigate beyond established models to comprehensively elucidate EPEC pathogenesis.
Keywords: GrlA; Ler; PerC; clinical isolates; enteropathogenic Escherichia coli; locus of enterocyte effacement; pathogenesis; phenotypic diversity; regulatory mechanisms; type III secretion system.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Levine MM, Nasrin D, Acácio S, Bassat Q, Powell H, et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob Health. 2020;8:e204–e214. doi: 10.1016/S2214-109X(19)30541-8. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

